We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox UC 11
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='1mnm' size='350' side='right' caption='YEAST MATALPHA2/MCM1/DNA TERNARY TRANSCRIPTION COMPLEX CRYSTAL STRUCTURE (PDB entry [[1mnm]])' scene=''> | <StructureSection load='1mnm' size='350' side='right' caption='YEAST MATALPHA2/MCM1/DNA TERNARY TRANSCRIPTION COMPLEX CRYSTAL STRUCTURE (PDB entry [[1mnm]])' scene=''> | ||
The structure of a complex containing the homeodomain repressor protein MATalpha2 and the MADS-box transcription factor '''MCM1''' bound to DNA has been determined by X-ray crystallography at 2.25 A resolution. | The structure of a complex containing the homeodomain repressor protein MATalpha2 and the MADS-box transcription factor '''MCM1''' bound to DNA has been determined by X-ray crystallography at 2.25 A resolution. | ||
| - | <Structure load='3pmq' size='400' frame='true' align='right' caption='MtrF General Structure containing 10 heme groups, pdb:[[3pmq]]' /> | ||
[[MtrF]] is a cell surface cytochrome on the Gram-negative bacteria known as ''Shewanella oneidensis''.<ref name="mtrf"> PMID:11418600</ref> MtrF is involved with shuttling electrons across its (''S. oneidensis'') outer surface. MtrF has several homologues, MtrC and the protein OmcA. These three different proteins are thought to be replaceable with one another in deletion mutation experiments.<ref name= "mtrf" /> | [[MtrF]] is a cell surface cytochrome on the Gram-negative bacteria known as ''Shewanella oneidensis''.<ref name="mtrf"> PMID:11418600</ref> MtrF is involved with shuttling electrons across its (''S. oneidensis'') outer surface. MtrF has several homologues, MtrC and the protein OmcA. These three different proteins are thought to be replaceable with one another in deletion mutation experiments.<ref name= "mtrf" /> | ||
__TOC__ | __TOC__ | ||
| - | |||
| - | ==MtrF Function== | ||
| - | |||
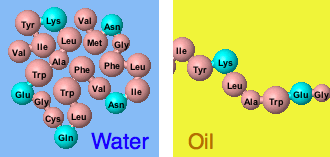
[[Image:MW_Folding_Simulations.gif]] | [[Image:MW_Folding_Simulations.gif]] | ||
| Line 13: | Line 9: | ||
<scene name='Sandbox_UC_11/Structura_secundaria/1'>TextToBeDisplayed</scene> | <scene name='Sandbox_UC_11/Structura_secundaria/1'>TextToBeDisplayed</scene> | ||
| + | ==Structure== | ||
Through the structure of this oligomer that was obtained by crystallography, we can see that HDAC9 is kept <scene name='Sandbox_Reserved_1/Binding/3'>relatively far from the genomic DNA </scene> even after MEF2 binding to the chromosome. We can also easily observe a very well <scene name='Sandbox_Reserved_1/Conservation/7'>evolutionary conserved region</scene> near the DNA binding site of MEF2. | Through the structure of this oligomer that was obtained by crystallography, we can see that HDAC9 is kept <scene name='Sandbox_Reserved_1/Binding/3'>relatively far from the genomic DNA </scene> even after MEF2 binding to the chromosome. We can also easily observe a very well <scene name='Sandbox_Reserved_1/Conservation/7'>evolutionary conserved region</scene> near the DNA binding site of MEF2. | ||
Current revision
| |||||||||||
Directed evolution and Colicin7/Immunity-proteins complexes[2]
| |||||||||||
- ↑ 1.0 1.1 Fotinou C, Emsley P, Black I, Ando H, Ishida H, Kiso M, Sinha KA, Fairweather NF, Isaacs NW. The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin. J Biol Chem. 2001 Aug 24;276(34):32274-81. Epub 2001 Jun 19. PMID:11418600 doi:10.1074/jbc.M103285200
- ↑ Levin KB, Dym O, Albeck S, Magdassi S, Keeble AH, Kleanthous C, Tawfik DS. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009 Oct;16(10):1049-55. Epub 2009 Sep 13. PMID:19749752 doi:10.1038/nsmb.1670
| |||||||||
| 1mnm, resolution 2.25Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Additional Literature
- Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat Struct Biol. 1999 Mar;6(3):243-52. PMID:10074943 doi:http://dx.doi.org/10.1038/6683
- Levin KB, Dym O, Albeck S, Magdassi S, Keeble AH, Kleanthous C, Tawfik DS. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009 Oct;16(10):1049-55. Epub 2009 Sep 13. PMID:19749752 doi:10.1038/nsmb.1670
External Resources
- Yahoo news coverage of luciferase studies funded research for military applications
- Luciferase at Wikipedia
- Luciferase: June 2006 Molecule of the Month as part of the series of tutorials that are at the RCSB Protein Data Bank and written by
- Lipase, the main article in Proteopedia.
- Molecular Playground/Pancreatic Lipase
- Lipase in Wikipedia