Sandbox Reserved 708
From Proteopedia
(→Structure) |
|||
| Line 17: | Line 17: | ||
As this diagram shows it the binary complex have β-sheets and α-helixes | As this diagram shows it the binary complex have β-sheets and α-helixes | ||
| - | This complex is composed by two domains A (green) and B (blue) linked by 12 hydrogen bonds in order to stabilize the molecule. But in this case only four are shown (with red dashed lines) with there <scene name='Sandbox_Reserved_708/Hbonds_amino_acids/7'>amino acids (17)</scene> engaged: Ser-66, Ser-215,Tyr-216, Arg-220, Gly-230, Asp-260, Gly-262, Val-263, Tyr-288, Ser-715,Tyr-716, Arg-720, Asp-760,Val-763, Asp-764, Tyr-788 and Glu-790 | + | This complex is composed by two domains A (green) and B (blue) linked by 12 hydrogen bonds in order to stabilize the molecule. But in this case only four are shown (with red dashed lines) with there <scene name='Sandbox_Reserved_708/Hbonds_amino_acids/7'>amino acids (17)</scene> engaged: Ser-66, Ser-215,Tyr-216, Arg-220, Gly-230, Asp-260, Gly-262, Val-263, Tyr-288, Ser-715,Tyr-716, Arg-720, Asp-760,Val-763, Asp-764, Tyr-788 and Glu-790. |
This structure also possesses <scene name='Sandbox_Reserved_708/Mg_ions/3'>two divalent cations Mg2+</scene> as well as <scene name='Sandbox_Reserved_708/Adp/3'>two molecules of ADP</scene>(Adenosine DiPhosphate) | This structure also possesses <scene name='Sandbox_Reserved_708/Mg_ions/3'>two divalent cations Mg2+</scene> as well as <scene name='Sandbox_Reserved_708/Adp/3'>two molecules of ADP</scene>(Adenosine DiPhosphate) | ||
These are the <scene name='Sandbox_Reserved_708/Catalytic_site/7'>amino acids</scene> engaged in the catalytic site of the protein, they are polar and localised in 3' end: Asp-181, Lys-183, Gln-185, Asn-186 and Ser-219. | These are the <scene name='Sandbox_Reserved_708/Catalytic_site/7'>amino acids</scene> engaged in the catalytic site of the protein, they are polar and localised in 3' end: Asp-181, Lys-183, Gln-185, Asn-186 and Ser-219. | ||
This enzyme is activated by phosphorylation at <scene name='Sandbox_Reserved_708/Tyr-216_phosphorylation/3'>Tyr-216</scene> and inactivated by phosphorylation at Ser-9 (not shown here because this structure start at residue 35). | This enzyme is activated by phosphorylation at <scene name='Sandbox_Reserved_708/Tyr-216_phosphorylation/3'>Tyr-216</scene> and inactivated by phosphorylation at Ser-9 (not shown here because this structure start at residue 35). | ||
| - | |||
| - | |||
Arg-141 is one of the key residues for <scene name='Sandbox_Reserved_708/Adp_recognition_by_arg141/1'>specific ATP/ADP recognition</scene> by TPK I/GSK3 . In this structure no residues are phosphorylated but the orientation of the activation loop in TPK I/GSK3 is similar to that in phosphorylated CDK2 and ERK2, suggesting that TPK I/GSK3 falls into a conformation that enables it to be constitutively active* <ref>Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 | Arg-141 is one of the key residues for <scene name='Sandbox_Reserved_708/Adp_recognition_by_arg141/1'>specific ATP/ADP recognition</scene> by TPK I/GSK3 . In this structure no residues are phosphorylated but the orientation of the activation loop in TPK I/GSK3 is similar to that in phosphorylated CDK2 and ERK2, suggesting that TPK I/GSK3 falls into a conformation that enables it to be constitutively active* <ref>Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 | ||
M. Aoki, T. Yokota, I. Sugiura, C. Sasaki, T. Hasegawa, C. Okumura, K. Ishiguro, T. Kohno, S. Sugio and T. Matsuzaki</ref>. | M. Aoki, T. Yokota, I. Sugiura, C. Sasaki, T. Hasegawa, C. Okumura, K. Ishiguro, T. Kohno, S. Sugio and T. Matsuzaki</ref>. | ||
Revision as of 18:02, 1 January 2013
Contents |
Descritpion
The Glycogen Synthase Kinase 3 beta- GSK3 beta, also known as the tau protein kinase I, is a serine protein kinase that participates in many different pathways regulating critical cellular functions as structure, gene expression, mobility, and apoptosis. GSK-3 phosphorylates a large number of substrates and is himself regulated by phosphorylation. GSK-3 is implicated in several diseases like Alzheimer’s disease, diabetes, mood disorders and cancer,[1]. In the Alzheimer’s disease, it is thought to be tied to the process of the hyperphosphorylation of tau proteins which leads to the formation of neurofibrillary tangles and to the build up of Amyloid-β (Aβ) deposits. Dephosphorylated tau binds normally to microtubules, one of the major components of the neuronal cytoskeleton that contributes to the proper function of neurons. Considering the roles plaid by GSK3 in promoting both pathological features of Alzheimer disease, GSK3-inhibitors may act positively in the therapy of Alzheimer’s patients. But due to the importance of its role in numerous cellular functions, it is important to develop inhibitors that do not affect or suppress its primary activity. There are two isoforms of GSK-3: GSK3α et GSK3β . Our concern focuses on GSK3 two mechanisms that affect the activity of the kinase are its inhibition by phosphorylation of serine-9 and its activity enhancement by phosphorylation of tyrosine-216.
Activity
Structure
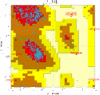 ,[2]This is the Ramachandran plot of 1j1c, here there is a link to understand this diagram.
As this diagram shows it the binary complex have β-sheets and α-helixes
,[2]This is the Ramachandran plot of 1j1c, here there is a link to understand this diagram.
As this diagram shows it the binary complex have β-sheets and α-helixes
This complex is composed by two domains A (green) and B (blue) linked by 12 hydrogen bonds in order to stabilize the molecule. But in this case only four are shown (with red dashed lines) with there engaged: Ser-66, Ser-215,Tyr-216, Arg-220, Gly-230, Asp-260, Gly-262, Val-263, Tyr-288, Ser-715,Tyr-716, Arg-720, Asp-760,Val-763, Asp-764, Tyr-788 and Glu-790. This structure also possesses as well as (Adenosine DiPhosphate) These are the engaged in the catalytic site of the protein, they are polar and localised in 3' end: Asp-181, Lys-183, Gln-185, Asn-186 and Ser-219. This enzyme is activated by phosphorylation at and inactivated by phosphorylation at Ser-9 (not shown here because this structure start at residue 35). Arg-141 is one of the key residues for by TPK I/GSK3 . In this structure no residues are phosphorylated but the orientation of the activation loop in TPK I/GSK3 is similar to that in phosphorylated CDK2 and ERK2, suggesting that TPK I/GSK3 falls into a conformation that enables it to be constitutively active* [3].
External Ressources
References
- ↑ Neurochem Res. 2007; 32(4-5): 577–595. Glycogen Synthase Kinase-3 (GSK3): Inflammation, Diseases, and Therapeutics, Richard S. Jope,* Christopher J. Yuskaitis, and Eléonore Beurel
- ↑ http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1j1c&template=procheck_summary.html
- ↑ Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 M. Aoki, T. Yokota, I. Sugiura, C. Sasaki, T. Hasegawa, C. Okumura, K. Ishiguro, T. Kohno, S. Sugio and T. Matsuzaki

