From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
|
| Line 41: |
Line 41: |
| | | | |
| | | | |
| - | <StructureSection Ligands and their Binding-SitesSection load='1s8o_mm1.pdb' size='400' side='left' caption='X-ray crystal structure of hsEH (PDB entry [[1s8o]])' scene='Sandbox_Reserved_714/Initial_scene/2'> | + | <StructureSection Ligands and their Binding-SitesSection load='1s8o_mm1.pdb' size='400' side='left' scene='Sandbox_Reserved_714/Initial_scene/2'> |
| | | | |
| | | | |
Revision as of 22:27, 1 January 2013
Template:Sandbox ESBS 2012
1OCY : ONE COMPONENT OF THE BACTERIOPHAGE T4 SHORT TAIL FIBRE
(STRUCTURE OF THE RECEPTOR-BINDING DOMAIN OF THE BACTERIOPHAGE T4 SHORT TAIL FIBRE)
Description of Bacteriophage T4
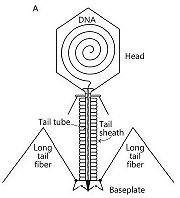
Schematic representation of bacteriophage T4
[1]Bacteriophage T4 belongs to the Myoviridae family and the Caudovirales order. It belongs to this order and family because of its complex tail structure. In this complex tail structures many proteins are involved. It infects Escherichia coli bacteria.
It consists of three parts : a DNA-containing head, a doubles-tubed tail with a contractile outer tail-sheath and a baseplate with long and short tail fibers.
Each bacteriophage T4 baseplate is composed of at least 16 different gene products, also called gp which are oligomeric proteins. These gene products can be divided in two groups: the six long and the six short tail fibers (on the schematic representation, on the left, they are not all shown). They form a multiprotein machine which plays an important role in the first stage of a phage infection. It is essential for the host cell recognition, the attachment of the bacteriophage and the sheath contraction allowing viral DNA ejection. [2]
Adsorption and penetration phases
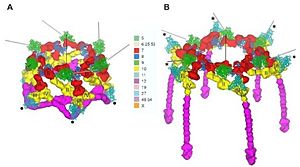
Deployement of short tails A: Hexagonal conformation, B: Star conformation gp12 in manganta and gp10 in yellow
[1]First, the viral particles recognize and bind reversibly to the outer membrane protein C (OmpC) or the cell-surface lipopolysaccharide receptors thanks to six long tail fibers which are connected to the baseplate. After at least three long tail fibers have bound, the baseplate conformation changes: from a hexagon shape, it becomes a six-pointed star. This change can be the result of changing the interactions between proteins. It has two consequences.
The first one is the unfolding of the short tail fibers, which are under the baseplate. Thus, they are able to attach irreversibly to the host cell surface.
The second one is the induction of the tail sheath’s contraction. Afterwards the tail tube is driving through the cell membrane. The activated lysozyme domain of gp5 degraded the peptidoglycan layer.
To finish, the phage DNA single-stranded is injected into the bacterial cytoplasm through the tail tube.
Presentation of gp12
During the first step of the lytic cycle, the short tail fibers gp12 attach the virion to the host cell.
Thanks to a three-dimensional cryoelectron microscopy, a reconstruction of the baseplate was determined to a resolution of 12 Angstrom. It has shown that the total length of the gp12 density is inferior at 340 Å. [1]
This technique was also a way to determine the interactions between the different proteins and their localization. Thus, interactions of the short tail fibers with each other and with the gp11 maintain the hexagon shape stability. The gp11 is also associated with gp10, which is clamped between the three fingers of gp11. This association between gp10 and gp11 is essential for the attachment of gp12 to the baseplate. When a rotation of gp11 around its three-fold axis occurs, the end of the short tail fiber is oriented toward the host cell surface.
After the creation of six wedge composed of gp11, gp10, gp7, gp8, gp6, gp25 and gp53, is the wedge associated to the hub. The hub is a complex of gp5-gp27. Then are gp9 and gp12 added. The chaperone protein gp57 help gp12 to adopt its correct folding into trimmers. To finish, gp48 and gp54 are added and it form the complete baseplate.
Structure [2]
Short tail fibres consist of the single protein . This protein forms a parallel, in-register, homo-trimer of 527 residues per subunit. 18 monomers are on the baseplate. 1ocy is a monomer of the short tail fibres. A monomer of gp12 has a mass of 55.3 kDa. The Gp12 N-terminal domain is bound to the baseplate. While the C-terminal globular domain is supposed to bind the bacterial host cell.
Gp12 can be divided into two fragments. One fragment with a mass of 33kDA and a second with a mass of 45kDa.
The 33kDa Fragment [3] [4]
The 33kDa fragment (PDB:1H6W) was generated in the presence of EDTA. This fragment contains the residues and . The residues 397-517 are lacking because of internal deletion.
The 33kDa fragment can be further sub-divided into two subunits. The (residue 333-341) and the collar (residues plus ). The neck connects the body of the fibre to its C-terminal collar and receptor binding-site. It consists of a triple alpha-helix which is built by the residues 333-341.
The collar domain is a small globular domain. It contains six beta-strands and an alpha-helix.
The 45kDa Fragment [2]
For generating the 45kDa fragment the full length gp12 was co-expressed with its chaperone gp57 and purified [1]. Like the 33kDa fragment it also starts with the amino acid Leu85. The 45kDa fragment contains the residues , which are in the 33kDa fragment internal deleted.
Like the 33kDa fragment the 45kDa fragment can also be divided into two subunits. These two subunits are called head (residues and residues ) and (residues 447-487). On the border between the head and the bonnet subunit there is a metal-binding site.
Receptor-Binding Domain [2]
Gp12 is fixed with its N-terminal domain to the baseplate. So the C-terminal domain has to be involved in LPS-binding. To detect where the receptor-binding domain is, full-length gp12, 33kDa fragments and 45kDa fragments were immobilised in micro-plate wells and were allowed to bind to bacteria [1]. The result was that the 33kDa fragment did never bind to a bacteria. The 45kDa fragment did bind. So the receptor-binding domain is absent in the 33kDa fragment but present in the 45kDa fragment. The residues which are present in the 45kDa fragment and lacking in the 33kDa fragment are the residues . They are referred to be part of the receptor-binding domain.
The receptor-binding domain can be sub-divided into head and bonnet. On the border between these two subdomains there is a metal-binding site. This site binds presumably to zinc. ( and ) from each monomere are octahedrally coordinated around the zinc.
The receptor-binding domain can be compared to a flower bud. This flower bud has got 12 petals which are organised in a 3-fold symmetry. At the bottom there are the residues 406-432 they form the , just above there are the residues 489-504 which form the . The is formed by the residues 450-470 and at the top there are the residues 470-480 and form the . The complete and active receptor-binding domain is built by the trimeric protein. This trimeric proteine structure is stabilised by many inter-domain hydrogen bounds. These hydrogen bounds can have the profiles: main-chain-main-chain, main-chain-side-chain and side-chain-side-chain.
The LPS-Bindind Site [2]
The exact position of the lipo-polysaccharide(LPS)-binding site is not known. Proteolysis experiments showed, that it contains to the domain with the residues . Compared with the homologous bacteriophage T4-like strain AR1 sequence, which also binds to the same LPS core molecule like gp12, there can be some possible binding residues be assumed.
:
, , ,
:
,, , , , ,, , ,
It can not be ruled out that also other amino acids are important for binding! Further it can not be ruled out that not all of these named amino acids are important for binding
The mentioned amino acids are starting point for site-directed mutations and /or other biochemical experiments to clarify the location of this binding site.
Ligands and their Binding-Sites
So4
In the gp12 there are two molecules of SO4 ligands. One of them interacts with the citric acid. The blue molecule is the SO4. The other . It builds two hydrogen bounds to Ser287 with the distances 2.66 and 3.20.
Citric Acid
The is drawn in green. It interacts with following amino acids: Arg465 (distance 3.01), Asp455(distance 2.42) and Tyr454 (distance2.94). . These interactions are caused by hydrogen bounds.
Zinc
The zinc is located in the centre of the receptor-binding domain. The zinc-binding site lies on the border between the head and the bonnet of the 45kDa domain. It interacts with the amino acids of each monomer. The distances of the zinc ion to the NE2 of and are 2.22 Å and 2.25 Å. The normally found distances between His and zinc are shorter. The explaination that these distances are longer than normally found is the octahedral coordination of the zinc in this structure.
The role of the zinc ion is probably absolute of structural nature. It increases the stability of the C-terminus of gp12 against proteases, but it also raises the stability of the C-terminus in general.
Applications
The most important role of gp12 is its ability to recognize and adhere to the host cell. Three other gene products (gp36, gp37 and gp38) are also targeting proteins and have a similar function. Nowadays some patents actually use random changes to create a bank of genetically modified bacteriophages. Recombinant bacteriophages are becoming a hope for treatment against multiresistant bacterial strains.
[5]
External Ressources
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Leiman PG, Arisaka F, van Raaij MJ, Kostyuchenko VA, Aksyuk AA, Kanamaru S, Rossmann MG. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010 Dec 3;7:355. doi: 10.1186/1743-422X-7-355. PMID:21129200 doi:10.1186/1743-422X-7-355
- ↑ 2.0 2.1 2.2 2.3 2.4 Kostyuchenko VA, Leiman PG, Chipman PR, Kanamaru S, van Raaij MJ, Arisaka F, Mesyanzhinov VV, Rossmann MG. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003 Sep;10(9):688-93. Epub 2003 Aug 17. PMID:12923574 doi:http://dx.doi.org/10.1038/nsb970
- ↑ Thomassen E, Gielen G, Schutz M, Schoehn G, Abrahams JP, Miller S, van Raaij MJ. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J Mol Biol. 2003 Aug 8;331(2):361-73. PMID:12888344
- ↑ van Raaij MJ, Schoehn G, Jaquinod M, Ashman K, Burda MR, Miller S. Identification and crystallisation of a heat- and protease-stable fragment of the bacteriophage T4 short tail fibre. Biol Chem. 2001 Jul;382(7):1049-55. PMID:11530935 doi:10.1515/BC.2001.131
- ↑ EP2097516 (B1) - Method for preparing bacteriophages modified by the insertion of random sequences in the screening proteins of said bacteriophages, IRIS FRANCOIS, PHERECYDES PHARMA
Proteopedia Page Contributors and Editors
Anne-Lise Terrier, Bianca Waßmer
| |



