Sandbox Reserved 714
From Proteopedia
(Difference between revisions)
| Line 38: | Line 38: | ||
9/10-phosphonoxy-hydroxy-octadecanoate + H<sub>2</sub>O ↔ 9/10-dihydroxy-octadecanoate + phosphate | 9/10-phosphonoxy-hydroxy-octadecanoate + H<sub>2</sub>O ↔ 9/10-dihydroxy-octadecanoate + phosphate | ||
| - | Its <scene name='Sandbox_Reserved_714/Nter_activesite/1'>active site</scene> contains several conserved aspartates in phosphatases and phosphonatases | + | Its <scene name='Sandbox_Reserved_714/Nter_activesite/1'>active site</scene> contains several conserved aspartates in phosphatases and phosphonatases: D9, D11, D184 and D185. This enzymatic activity is Mg<sup>2+</sup> dependant, because the structure of the active site is in its optimal conformation when the cation makes coordination interactions. When the catalytic activity of the N-term domain is available, Magnesium is octahedrally coordinated with the four aspartates, one water molecule and the phosphate belonging to the substrate. We can note that Mg<sup>2+</sup> is not directly involved in the catalytic mechanism, and all its interactions with the active site remain during the hydrolysis. Its single role consists in maintaining the three-dimensional structure of the active site. |
First, the oxygen on the lateral chain of D9 attacks the phosphate. After the addition of a proton H<sub>+</sub>, the product with its two hydroxyl functions is released, while the phosphate is still linked to D9. Then, a waters molecule binds the phosphate, breaking its bond with the aspartate. Therefore the phosphate can be finally released and the active site can accept a lipid again and start a new catalytic cycle. | First, the oxygen on the lateral chain of D9 attacks the phosphate. After the addition of a proton H<sub>+</sub>, the product with its two hydroxyl functions is released, while the phosphate is still linked to D9. Then, a waters molecule binds the phosphate, breaking its bond with the aspartate. Therefore the phosphate can be finally released and the active site can accept a lipid again and start a new catalytic cycle. | ||
Revision as of 19:48, 2 January 2013
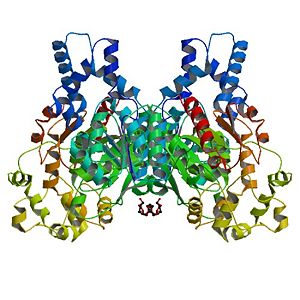
Human Soluble Epoxide Hydrolase: Biological assembly, 1s8o
Contents |
Overview
| |||||||||||
Additional 3D Structures of hsEH
1vj5 - hsEH + N-cyclohexyl-N'-(4-iodophenyl)urea complex
1zd2,1zd3, 1zd4,1zd5 - hsEH + 4-(3-cyclohexyluriedo)-carboxylic acids
3ant - Hydrolase domain + synthetic inhibitor
3pdc - Hydrolase domain + benzoxazole inhibitor
External ressources
Protein Data Bank entry on 1S8O
Uniprot link on Bifunctional epoxyde hydrolase 2
Wikipedia page on Epoxyde hydrolase 2
References
- ↑ Morisseau C, Hammock BD. Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol. 2005;45:311-33. PMID:15822179 doi:10.1146/annurev.pharmtox.45.120403.095920
- ↑ Gomez GA, Morisseau C, Hammock BD, Christianson DW. Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis. Biochemistry. 2004 Apr 27;43(16):4716-23. PMID:15096040 doi:10.1021/bi036189j
- ↑ Newman JW, Morisseau C, Harris TR, Hammock BD. The soluble epoxide hydrolase encoded by EPXH2 is a bifunctional enzyme with novel lipid phosphate phosphatase activity. Proc Natl Acad Sci U S A. 2003 Feb 18;100(4):1558-63. Epub 2003 Feb 6. PMID:12574510 doi:10.1073/pnas.0437724100
Proteopedia Page Contributors and Editors
DUTREUX Fabien, BONHOURE Anna

