Sandbox Reserved 713
From Proteopedia
| Line 47: | Line 47: | ||
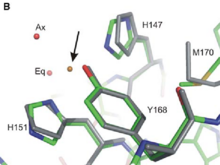
Thanks to its extracellular Cu-Binding Domain (CuBD) constituated by the amino acids describe above, APP can modulate '''copper transport and storage'''. The Cu-binding domain of the apo protein (show in grey in the fig 1 ) seems to be able to fixe '''Cu(II) and to reduce it into Cu(I)'''. More precisely, the Tyr 168, the His 147 and the His 151 participate in Cu(II) binding but not the Methionine 170. In addition, two molecules of water, one axial and one equatorial (noted Ax and Eq on Fig 1) play an important role in the formation of the APP-Cu(II)complex (represented in standard atomic colouring in Fig 1). (PDB ID : [http://www.rcsb.org/pdb/explore.do?structureId=2fk1 2fk1]). It results that the arrangement of the atoms involved in the Cu(II) binding adopts a square pyramidal geometry which can be classified as a Type 2 non-blueCu(II) center. In this type of center Cu(II) is bound by two or three nitrogene ligands and one or two oxygen ligands. Met170 is supposed to act as an electron donor to Cu(II). | Thanks to its extracellular Cu-Binding Domain (CuBD) constituated by the amino acids describe above, APP can modulate '''copper transport and storage'''. The Cu-binding domain of the apo protein (show in grey in the fig 1 ) seems to be able to fixe '''Cu(II) and to reduce it into Cu(I)'''. More precisely, the Tyr 168, the His 147 and the His 151 participate in Cu(II) binding but not the Methionine 170. In addition, two molecules of water, one axial and one equatorial (noted Ax and Eq on Fig 1) play an important role in the formation of the APP-Cu(II)complex (represented in standard atomic colouring in Fig 1). (PDB ID : [http://www.rcsb.org/pdb/explore.do?structureId=2fk1 2fk1]). It results that the arrangement of the atoms involved in the Cu(II) binding adopts a square pyramidal geometry which can be classified as a Type 2 non-blueCu(II) center. In this type of center Cu(II) is bound by two or three nitrogene ligands and one or two oxygen ligands. Met170 is supposed to act as an electron donor to Cu(II). | ||
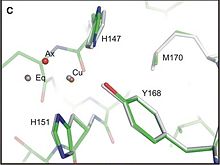
| - | In the Cu(I) binding geometry (represented in grey in Fig 2)(PDB id [http://www.rcsb.org/pdb/explore.do?structureId=2fk2 2fk2]), there is no axial water molecules ; the site adopts also a distorted square planar arrangement which is unfavorable for Cu(I) suggesting that this states is not favorable and can lead to the tranfert of the Cu (I) to others proteins.In Fig 2 standart atomic colouring reprensents the Cu(II)binding form. | + | In the Cu(I) binding geometry (represented in grey in Fig 2)(PDB id [http://www.rcsb.org/pdb/explore.do?structureId=2fk2 2fk2]), there is no axial water molecules ; the site adopts also a distorted square planar arrangement which is unfavorable for Cu(I) suggesting that this states is not favorable and can lead to the tranfert of the Cu (I) to others proteins.In Fig 2 standart atomic colouring reprensents the Cu(II)binding form.</StructureSection> |
=='''Medical Implication'''== | =='''Medical Implication'''== | ||
| Line 59: | Line 59: | ||
If the CuBD (located in E1 region) does contribute to APP dimerisation without Cu ions, that mean that the Cu binding may reduce AB production, either by shifting the monomer-dimer equilibrium to favor monomer form, or by re-orienting the dimer form, which disfavour the aB production. <ref> PMID : 18030462 <ref/> | If the CuBD (located in E1 region) does contribute to APP dimerisation without Cu ions, that mean that the Cu binding may reduce AB production, either by shifting the monomer-dimer equilibrium to favor monomer form, or by re-orienting the dimer form, which disfavour the aB production. <ref> PMID : 18030462 <ref/> | ||
!!!!!!!!!!!!!!!! | !!!!!!!!!!!!!!!! | ||
| - | + | ||
=='''Additionnal Resources'''== | =='''Additionnal Resources'''== | ||
Revision as of 12:14, 5 January 2013
Alzheimer's amyloid precursor protein copper-binding domain
| |||||||||
| 2fkl, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: | APP (Homo sapiens) | ||||||||
| Related: | 1owt, 2fjz, 2fk1, 2fk2, 2fk3 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Introduction
2FKL is located in a transmembrane protein called APP for Amyloid precursor protein. This domain going from residue 124 to 189 is localized in the extracellular part of APP just between the Growth Factor Domain (GFD) and the Acidic domain, in a region called Cu-Binding Domain which is able to bind Cu and Zinc.
This proteins plays a major role into the development of Alzheimer disease[1]. APP cleavage by BACE and gamma secretase gives ended rise to the Aβ peptide, which forms at the end an aggregation of amyloid plaques [2] .
As the interaction between copper ion and APP can modulate the production of Aβ peptide [3] and also the progression of Alzheimer disease, structural studies of the Cu-binding Domain of this protein give a lot of information for the development of novel therapeutics.
Structure
| |||||||||||
Medical Implication
An Alzheimer disease is responsible for the most comon form of dementia among old people.)Causes of this disease are not clear but genes and environmental factors seem to play a major role. (lien au dessus) It is characterized by extensive neuronal death, shrinkage of the brain and also the presence of amyloid plaques into the brain.( article principale) These plaques are concentrated with Aβ peptide which results from the anormal cleavage of APP by successive action of the β and γ secretases. The alteration in copper homostasis is directly related to the progression of Alzheimer disease. The impact of Cu(II) on the aggregation of Aβ peptide is still discussed even if Cu(II) is find in elevated concentration in these plaques. [6]
If the CuBD (located in E1 region) does contribute to APP dimerisation without Cu ions, that mean that the Cu binding may reduce AB production, either by shifting the monomer-dimer equilibrium to favor monomer form, or by re-orienting the dimer form, which disfavour the aB production. [7]