Sandbox Reserved 706
From Proteopedia
(→'''Structure of the complex''') |
|||
| Line 83: | Line 83: | ||
-<scene name='Sandbox_Reserved_706/Phe81_leu84_phe88/1'>Phe81, Leu84, and Phe88</scene> from <scene name='Sandbox_Reserved_706/Switch2_rab27b/1'>switch 2</scene> of Rab27B participate in {{Template:ColorKey_Hydrophobic}} interactions with the residues from <scene name='Sandbox_Reserved_706/Alpha_5_et_1_de_slac2/1'>α1 and α5</scene> of Slac2-a. | -<scene name='Sandbox_Reserved_706/Phe81_leu84_phe88/1'>Phe81, Leu84, and Phe88</scene> from <scene name='Sandbox_Reserved_706/Switch2_rab27b/1'>switch 2</scene> of Rab27B participate in {{Template:ColorKey_Hydrophobic}} interactions with the residues from <scene name='Sandbox_Reserved_706/Alpha_5_et_1_de_slac2/1'>α1 and α5</scene> of Slac2-a. | ||
| - | Furthermore, Trp73 of Rab27B is located in the middle of this contact area, and it plays a central role in {{Template:ColorKey_Hydrophobic}} interactions but also for Slac2-a binding. .<ref>PMID:18940604</ref> | + | Furthermore, <scene name='Sandbox_Reserved_706/Try73/1'>Trp73 of Rab27B</scene> is located in the middle of this contact area, and it plays a central role in {{Template:ColorKey_Hydrophobic}} interactions but also for Slac2-a binding. .<ref>PMID:18940604</ref> |
-There are also electrostatic interactions involved in this Slac2-a binding, for instance amino acids Glu14, Val18, and Val21 of Slac2-a are required for this interaction. | -There are also electrostatic interactions involved in this Slac2-a binding, for instance amino acids Glu14, Val18, and Val21 of Slac2-a are required for this interaction. | ||
Revision as of 11:39, 6 January 2013
Please, do not delete or modify text or images, this page is reserved for a work from two students in ESBS. Thank you.
Contents |
Structure
2ZET is a 4 chains structure of sequences from Mus musculus. Full crystallographic information is available from OCA.
Structure of Rab27B
General informations:
To obtain structural information on the complex of mouse Rab27B and Slac-a, a C-terminally truncated form of the GTPase-deficient mutant Rab27B (Q78L) was used. In fact, full size of Rab27B is 218 amino acids long whereas the truncated form of Rab27B contains only the GTPase domain (residues 1 to 201). [1] [2]
Detailed structure:
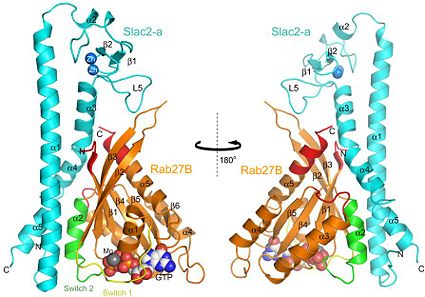
The structure of Rab27B in the complex contains a (ß1-6) flanked by (α1–5). Like other Ras-like small GTPases, Rab27B binds with and in the conserved nucleotide-binding site (also see figure 2). Rab27B adopt a globular form in the complex. The and of Rab27B are both regions involved in Slac2-a binding. [3]
The complex Rab27B/Slac2-a can only be formed when Rab27B is in a GTP form. This can be explained because of the changing of shape between Rab27B in the GTP and GDP form. (See figure 3)
Rab27B–GTP adopts a compact monomer conformation, while an extended conformation was observed for GDP-bound Rab27B.
and of Rab27B play an important role in this changing of conformation. In fact, switch 1 and 2 allow GTP binding by making hydrogen bond with GTP's γ-phosphate, and this way also allow Slac2-a binding. But in the complex Rab27B/GDP switch 1 and switch 2 exist completely apart from the bound GDP, so that there are no interactions with Slac2-a and no binding.
Strucure of Slac2-a
General informations:
Slac2-a is a fragment from Slp Homology Domain, residues 1-146 from Melanophilin molecule. This protein is also called Exophilin-3. [4] Slac2-a plays an important role in actin-based melanosome transport in mammalian. In fact, the actin-binding domain of Slac2-a/Melanophilin is required for melanosome distribution in melanocytes.[5]
To obtain structural information on the complex of mouse Rab27B/Slac2-a, a truncated form of Slac2-a was used. This fragment of Slac2-a is the minimum region (residue 1 to 146) of Slac2-a that specifically binds to the GTP-bound form of Rab27B.
Detailed structure:
The structure of the Slac2-a effector domain comprises three subdomains: SHD1, SHD2, and a zinc-binding subdomain that is flanked by the two SHDs. [6] SHD1 is folded into a 66 Å , and SHD2 is composed of . SHD1 and SHD2 interact with each other by making a coiled coil between α1 and α3–α5. In this coiled coil, α3 and α5 interact with α1 in an antiparallel manner (see figure 2). The contains four short Beta Strands (ß1–ß4) and five loops(L1-L5), including one short Alpha Helices α2 and three short helices (in pupule). The are each coordinated by four conserved cysteine residues: for Zn1 binding, and for Zn2 binding. [7]
|
Structure of the complex
Model and general structure :
The final model of 2ZET includes:
- two pairs of the Rab27B/Slac2-a complex ,called and complex (A and B designate Rab 27B and C and D designate Slac2-a),
- ,
- ,
- ,
- .
Because the structures of the two Rab27B/Slac2-a complex are the same we will describe only the structure of A/C complex with for ligand one GTP molecule, one magnesium ion, two zinc ions and one sulfate ion.
The A/C complex is a 3.0 Å crystal structure of the effector domain of mouse Slac2-a complexed with a GTPase-deficient mutant (Q78L) of mouse Rab27B. Rab27B adopt a globular form in the complex and Slac2-a forms an elongated helical fold with a subdomain that contain two zinc ions.
Interface of Rab27B/Slac2-a and interaction details:
Rab27B interacts with Slac2-a with a buried surface area of 2534 Ų, which corresponds to 30.2% of the total surface area. Rab27B binds to Slac2-a with good shape complementarity and this interaction involves many hydrophobic interactions and several electrostatic interactions. There are three contact areas in the interface between Rab27B and Slac2-a. The first and second contact areas involve the coiled-coil regions of Slac2-a and constitute the main interface.
- The first contact area of the Rab27B/Slac2-a interface consists of , part of the switch and interswitch regions of Rab27B, and the coiled-coil regions of Slac2-a.[8] In this first contact area we can find three different types of interactions, electrostatic, hydrophobic and hydrogen bonds: (see figure 3A)
- from of Rab27B participate in Hydrophobic interactions with the residues from of Slac2-a. Furthermore, is located in the middle of this contact area, and it plays a central role in Hydrophobic interactions but also for Slac2-a binding. .[9]
-There are also electrostatic interactions involved in this Slac2-a binding, for instance amino acids Glu14, Val18, and Val21 of Slac2-a are required for this interaction. Arg35 of Slac2-a is also engaged in an electrostatic interaction with the Asp7 carbonyl group of Rab27B .[10]
-In the first contact area, hydrogen bonds between Rab27B/Slac2-a were also observed. First, Asp91 of Rab27B, which confers a negative charge in the center of the interface with Slac2-a, forms hydrogen bonds with Arg29 and Gly133 of Slac2-a. Secondly, . Third, Arg90 of Rab27B forms a hydrogen bond with the Arg131 carbonyl group from the α4–α5 loop of Slac2-a. To finish, Glu32 of Slac2-a forms a hydrogen bond with Tyr6 of Rab27B. [11]
- The second contact area is formed by the Rab complementarity-determining regions (RabCDRs) of Rab27B and 117-SLEWYY-122 motif of Slac2-a helical region (also see Figure 6C). [12] [13]
At this interface, there are hydrophobic interactions and three hydrogen bonds. Tyr121 of Slac2-a form a hydrogen bond with the Arg90 carbonyl group of Rab27B. Arg128 of Slac2-a, which is highly conserved among the Slp-family proteins, makes two hydrogen bonds with the carbonyl groups of Gln118 and Ala121 of Rab27B
- The third contact area is the smallest one . It involves the zinc-binding subdomain of Slac2-a and the b2–b3 loop of Rab27B, with superposition of their molecular surfaces. (See figure 3E)
These findings strongly suggested that Tyr6 from RabCDR and three residues (Leu84, Phe88, and Asp91) from switch 2 of Rab27 are the minimum determinants of Slac2-a binding.[14]
External Resources
References
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ http://oca.weizmann.ac.il/oca-bin/ocaids?id=2zet
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ http://oca.weizmann.ac.il/oca-bin/ocaids?id=2zet
- ↑ Kuroda TS, Ariga H, Fukuda M. The actin-binding domain of Slac2-a/melanophilin is required for melanosome distribution in melanocytes. Mol Cell Biol. 2003 Aug;23(15):5245-55. PMID:12861011
- ↑ Fukuda M, Kuroda TS, Mikoshiba K. Slac2-a/melanophilin, the missing link between Rab27 and myosin Va: implications of a tripartite protein complex for melanosome transport. J Biol Chem. 2002 Apr 5;277(14):12432-6. Epub 2002 Feb 20. PMID:11856727 doi:10.1074/jbc.C200005200
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999 Feb 5;96(3):363-74. PMID:10025402
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014
- ↑ Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure. 2008 Oct 8;16(10):1478-90. PMID:18940604 doi:10.1016/j.str.2008.07.014