1ybq
From Proteopedia
(New page: 200px<br /><applet load="1ybq" size="450" color="white" frame="true" align="right" spinBox="true" caption="1ybq, resolution 2.0Å" /> '''Crystal structure of ...) |
|||
| Line 1: | Line 1: | ||
| - | [[Image:1ybq.jpg|left|200px]]<br /><applet load="1ybq" size=" | + | [[Image:1ybq.jpg|left|200px]]<br /><applet load="1ybq" size="350" color="white" frame="true" align="right" spinBox="true" |
caption="1ybq, resolution 2.0Å" /> | caption="1ybq, resolution 2.0Å" /> | ||
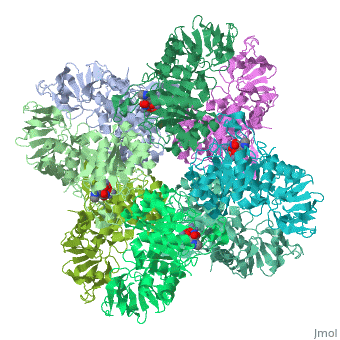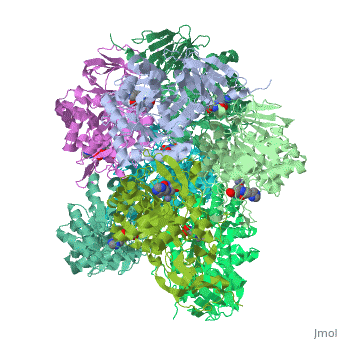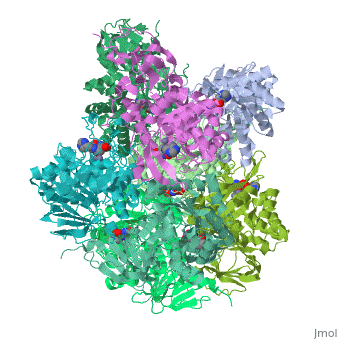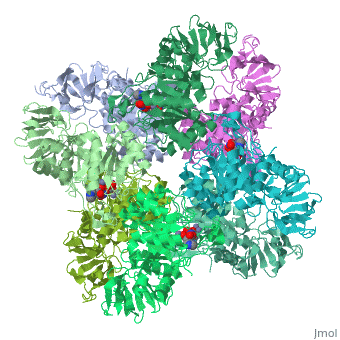
'''Crystal structure of Escherichia coli isoaspartyl dipeptidase mutant D285N complexed with beta-aspartylhistidine'''<br /> | '''Crystal structure of Escherichia coli isoaspartyl dipeptidase mutant D285N complexed with beta-aspartylhistidine'''<br /> | ||
==Overview== | ==Overview== | ||
| - | Isoaspartyl dipeptidase (IAD) is a member of the amidohydrolase | + | Isoaspartyl dipeptidase (IAD) is a member of the amidohydrolase superfamily and catalyzes the hydrolytic cleavage of beta-aspartyl dipeptides. Structural studies of the wild-type enzyme have demonstrated that the active site consists of a binuclear metal center positioned at the C-terminal end of a (beta/alpha)(8)-barrel domain. Steady-state kinetic parameters for the hydrolysis of beta-aspartyl dipeptides were obtained at pH 8.1. The pH-rate profiles for the hydrolysis of beta-Asp-Leu were obtained for the Zn/Zn-, Co/Co-, Ni/Ni-, and Cd/Cd-substituted forms of IAD. Bell-shaped profiles were observed for k(cat) and k(cat)/K(m) as a function of pH for all four metal-substituted forms. The pK(a) of the group that must be unprotonated for catalytic activity varied according to the specific metal ion bound in the active site, whereas the pK(a) of the group that must be protonated for catalytic activity was relatively independent of the specific metal ion present. The identity of the group that must be unprotonated for catalytic activity was consistent with the hydroxide that bridges the two divalent cations of the binuclear metal center. The identity of the group that must be protonated for activity was consistent with the free alpha-amino group of the dipeptide substrate. Kinetic constants were obtained for the mutant enzymes at conserved residues Glu77, Tyr137, Arg169, Arg233, Asp285, and Ser289. The catalytic properties of the wild-type and mutant enzymes, coupled with the X-ray crystal structure of the D285N mutant complexed with beta-Asp-His, are consistent with a chemical reaction mechanism for the hydrolysis of dipeptides that is initiated by the polarization of the amide bond via complexation to the beta-metal ion of the binuclear metal center. Nucleophilic attack by the bridging hydroxide is facilitated by abstraction of its proton by the side chain carboxylate of Asp285. Collapse of the tetrahedral intermediate and cleavage of the carbon-nitrogen bond occur with donation of a proton from the protonated form of Asp285. |
==About this Structure== | ==About this Structure== | ||
| - | 1YBQ is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli] with ZN and BDH as [http://en.wikipedia.org/wiki/ligands ligands]. Full crystallographic information is available from [http:// | + | 1YBQ is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli] with <scene name='pdbligand=ZN:'>ZN</scene> and <scene name='pdbligand=BDH:'>BDH</scene> as [http://en.wikipedia.org/wiki/ligands ligands]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1YBQ OCA]. |
==Reference== | ==Reference== | ||
| Line 13: | Line 13: | ||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: Protein complex]] | [[Category: Protein complex]] | ||
| - | [[Category: Davis, M | + | [[Category: Davis, M L.]] |
[[Category: Fresquet, V.]] | [[Category: Fresquet, V.]] | ||
| - | [[Category: Holden, H | + | [[Category: Holden, H M.]] |
[[Category: Marti-Arbona, R.]] | [[Category: Marti-Arbona, R.]] | ||
| - | [[Category: Raushel, F | + | [[Category: Raushel, F M.]] |
| - | [[Category: Thoden, J | + | [[Category: Thoden, J B.]] |
[[Category: BDH]] | [[Category: BDH]] | ||
[[Category: ZN]] | [[Category: ZN]] | ||
| Line 24: | Line 24: | ||
[[Category: hydrolase]] | [[Category: hydrolase]] | ||
| - | ''Page seeded by [http:// | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 16:03:47 2008'' |
Revision as of 14:03, 21 February 2008
|
Crystal structure of Escherichia coli isoaspartyl dipeptidase mutant D285N complexed with beta-aspartylhistidine
Overview
Isoaspartyl dipeptidase (IAD) is a member of the amidohydrolase superfamily and catalyzes the hydrolytic cleavage of beta-aspartyl dipeptides. Structural studies of the wild-type enzyme have demonstrated that the active site consists of a binuclear metal center positioned at the C-terminal end of a (beta/alpha)(8)-barrel domain. Steady-state kinetic parameters for the hydrolysis of beta-aspartyl dipeptides were obtained at pH 8.1. The pH-rate profiles for the hydrolysis of beta-Asp-Leu were obtained for the Zn/Zn-, Co/Co-, Ni/Ni-, and Cd/Cd-substituted forms of IAD. Bell-shaped profiles were observed for k(cat) and k(cat)/K(m) as a function of pH for all four metal-substituted forms. The pK(a) of the group that must be unprotonated for catalytic activity varied according to the specific metal ion bound in the active site, whereas the pK(a) of the group that must be protonated for catalytic activity was relatively independent of the specific metal ion present. The identity of the group that must be unprotonated for catalytic activity was consistent with the hydroxide that bridges the two divalent cations of the binuclear metal center. The identity of the group that must be protonated for activity was consistent with the free alpha-amino group of the dipeptide substrate. Kinetic constants were obtained for the mutant enzymes at conserved residues Glu77, Tyr137, Arg169, Arg233, Asp285, and Ser289. The catalytic properties of the wild-type and mutant enzymes, coupled with the X-ray crystal structure of the D285N mutant complexed with beta-Asp-His, are consistent with a chemical reaction mechanism for the hydrolysis of dipeptides that is initiated by the polarization of the amide bond via complexation to the beta-metal ion of the binuclear metal center. Nucleophilic attack by the bridging hydroxide is facilitated by abstraction of its proton by the side chain carboxylate of Asp285. Collapse of the tetrahedral intermediate and cleavage of the carbon-nitrogen bond occur with donation of a proton from the protonated form of Asp285.
About this Structure
1YBQ is a Protein complex structure of sequences from Escherichia coli with and as ligands. Full crystallographic information is available from OCA.
Reference
Mechanism of the reaction catalyzed by isoaspartyl dipeptidase from Escherichia coli., Marti-Arbona R, Fresquet V, Thoden JB, Davis ML, Holden HM, Raushel FM, Biochemistry. 2005 May 17;44(19):7115-24. PMID:15882050
Page seeded by OCA on Thu Feb 21 16:03:47 2008