Lactate Dehydrogenase
From Proteopedia
| Line 20: | Line 20: | ||
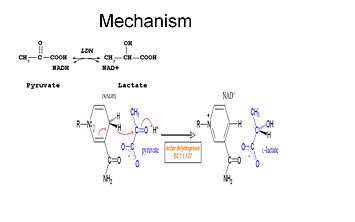
Studies have shown that the reaction mechanism of LDH follows an ordered sequence. [[Image:LDH_reaction.jpeg|355px|Catalytic function of LDH (1)]] | Studies have shown that the reaction mechanism of LDH follows an ordered sequence. [[Image:LDH_reaction.jpeg|355px|Catalytic function of LDH (1)]] | ||
{{Clear}} | {{Clear}} | ||
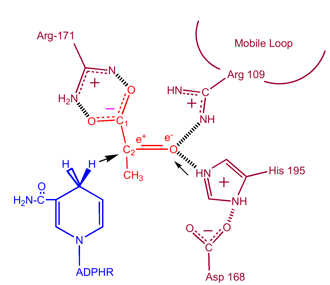
| - | In order for lactate to be oxidized NADH must bind to the enzyme first followed by lactate. Several residues are involved in the binding of NADH | + | In order for lactate to be oxidized NADH must bind to the enzyme first followed by lactate. <scene name='Lactate_Dehydrogenase/Secondary/5'>Several residues are involved in the binding of NADH</scene>. Once the NADH is bound to the enzyme, it is then possible for lactate to bind (substrate oxamate is shown; the ‑CH3 group is replaced by ‑NH2 to form oxamate). Lactate binds to the enzyme between the nicotinamide ring and several LDH residues. Transfer of a hydride ion then happens quickly in either direction giving a mixture of the two tertiary complexes, enzyme-NAD+-lactate and enzyme-NADH-pyruvate .Finally pyruvate dissociates from the enzyme followed by NAD+<ref name="1st"> http://www.bioc.aecom.yu.edu/labs/calllab/highlights/LDH.htm</ref>. |
[[Image:2nd.png|355px|(1)]] | [[Image:2nd.png|355px|(1)]] | ||
Revision as of 15:19, 10 January 2013
| |||||||||||
Contents |
3D structures of lactate dehydrogenase
Updated June 2012
2e37, 2v6m – TtL-LDH – Thermus thermophilus
2xxe, 3zzn, 4a73– TtL-LDH (mutant)
2x8l, 1ceq – PfL-LDH – Plasmodium falciparum
2zqy, 2zqz, 1llc – LcL-LDH – Lactobacillus casei
3d0o - SaL-LDH – Staphylococcus aureus
2v65 - L-LDH A chain – Champsocephalus gunnari
2v6b – L-LDH – Deinococcus radiodurans
2frm – CpLDH – Cryptosporidium parvum
1sov, 1pze - TgL-LDH – Toxoplasma gondii
1v6a – L-LDH A chain – Cyprinus carpio
1y6j – L-LDH – Clostridium thermocellum
1ldb - GsL-LDH – Geobacillus stearothermophilus
1ldm, 6ldh, 8ldh – sdM4-LDH – spiny dogfish
2ldx – C4-LDH - mouse
3pqe – BsL-LDH (mutant) – Bacillus subtilis
L-LDH from yeast (cytochrome b2)
1kbj - LDH FMN-binding domain
1sze, 1lco, 2oz0 - LDH FMN-binding domain (mutant)
1kbi - LDH FMN-binding domain + pyruvate
1szf, 1ldc - LDH FMN-binding domain (mutant) + pyruvate
1szg - LDH FMN-binding domain + sulfo-FMN
3ks0 - LDH residues 86-180 + flavocytochrome b2 heme domain + FAB B2B4
L-LDH binary complex
2ydn – PfL-LDH + bicine
1cet - PfL-LDH + chloroquine
1t2c - PfL-LDH + NAD
1ez4 - L-LDH + NAD – Lactobacillus pentosus
1lld, 2ldb - GsL-LDH + NAD
2a92 – PvL-LDH + NAD – Plasmodium vivax
2a94, 2aa3 - PvL-LDH + NAD-analog
1u4o, 1u4s, 1u5a, 1xiv – PfL-LDH + inhibitor
2xxb – TtL-LDH (mutant) + AMP
3pqf – BsL-LDH (mutant) + NAD
3vkv - LcLDH + fructose-bisphosphate
4aj1, 4aj2, 4aj4, 4aje, 4ajh, 4aji, 4ajj, 4ajk, 4ajl, 4ajn, 4ajo, 4al4 – LDH α chain + inhibitor – rat
4ajp – hLDH α chain + inhibitor - human
L-LDH ternary complex with inhibitor
1u5c - PfL-LDH + inhibitor + NAD
1t24, 1t25, 1t26 - PfL-LDH + azole derivative + NAD
1t2e - PfL-LDH (mutant) + oxamate + NAD
1ldg - PfL-LDH + oxamate + NAD
2xxj – TtL-LDH (mutant) + oxamate + NAD
2v7p - TtL-LDH + oxamate + NAD
3om9, 3czm – TgL-LDH + OXQ + NAD
3h3f – L-LDH A chain + oxamate + NAD – rabbit
2fnz - CpLDH + oxamate + NAD
1t2f - hL-LDH B chain + azole derivative + NAD
1i0z - hL-LDH H chain + oxamate + NAD
1i10 - hL-LDH M chain + oxamate + NAD
1oc4 – L-LDH + oxamate + NAD – Plasmodium berghei
1a5z - L-LDH + oxamate + NAD – Thermotoga maritima
1lth - L-LDH + oxamate + NAD – Bifidobacterium longum
1ldn - GsL-LDH + oxamate + NAD
9ldb, 9ldt - pL-LDH + oxamate + NAD – pig
L-LDH ternary complex with reactants
1sow, 1pzh - TgL-LDH + oxalate + NAD
1t2d - PfL-LDH + oxalate + NAD
1pzf - TgL-LDH + oxalate + NAD-analog
1pzg - TgL-LDH + sulfate + NAD-analog
3h3j – SaL-LDH (mutant) + pyruvate + NAD
3d4p - SaL-LDH + pyruvate + NAD
2fn7 - CpLDH + lactate + NAD
2ewd - CpLDH + pyruvate + NAD-analog
2fm3 - CpLDH + pyruvate + NAD
5ldh - pLDH + citrate + NAD-analog
3ldh - sdLDH + pyruvate + NAD
3pqd - BsLDH + fructose-bisphosphate + NAD
D-LDH
3kb6 – D-LDH + lactate + NAD – Aquifex aeolicus
1j49 – LdD-LDH + sulfate + NAD – Lactobacillus delbrueckii
1j4a - LdD-LDH + sulfate
1f0x – D-LDH + FAD – Escherichia coli
2dld - D-LDH + oxamate + NAD – Lactobacillus helveticus
L-LDH II
L-Lactate/malate dehydrogenase
1hye, 1hyg – L-LDH/MDH – Methanocaldococcus jannaschi
Additional Information
For additional information, see Carbohydrate Metabolism
Reference
- 1- http://www.bioc.aecom.yu.edu/labs/calllab/highlights/LDH.htm
- 2- http://www.cheric.org/ippage/e/ipdata/2004/05/file/e200405-701.pdf
- 3- http://resources.metapress.com/pdf-preview.axd?code=ulnhp23038060m21&size=largest
- 4- http://www.u676.org/Documents/Chretien-ClinChimActa-95.pdf
- 5- http://www.jbc.org/content/243/17/4526.full.pdf+html
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Ann Taylor, Jasper Small, David Canner