Group:MUZIC:Obscurin
From Proteopedia

| Line 7: | Line 7: | ||
The aminoacid sequences of human obscurin are deposited in the UniProt database (see <span style= | The aminoacid sequences of human obscurin are deposited in the UniProt database (see <span style= | ||
"font-variant:small-caps"> '''Uniprot'''</span> for [http://www.uniprot.org/uniprot/Q5VST9 ]) and GenBank GenBank ([http://www.ncbi.nlm.nih.gov/gene/84033 OBSCN obscurin]). The sequence provided in Figure 1 is based on the analysis of the human splice variants <ref>PMID: 16625312</ref>. Based on these different splice pathways, at least 4 isoforms are predicted: 2 long isoforms, two shorter isoforms in the extremely C-terminal part are possible but currently uncharacterized. | "font-variant:small-caps"> '''Uniprot'''</span> for [http://www.uniprot.org/uniprot/Q5VST9 ]) and GenBank GenBank ([http://www.ncbi.nlm.nih.gov/gene/84033 OBSCN obscurin]). The sequence provided in Figure 1 is based on the analysis of the human splice variants <ref>PMID: 16625312</ref>. Based on these different splice pathways, at least 4 isoforms are predicted: 2 long isoforms, two shorter isoforms in the extremely C-terminal part are possible but currently uncharacterized. | ||
| - | |||
| - | |||
| - | <Structure load='2wp3' size='300' side='right' caption='Crystal Structure of the titin Ig-like domain Complex between the titin M10 (blue) - Obscurin like 1 (Red) (PDB entry: [http://www.pdb.org/pdb/explore/explore.do?structureId=2wp3 2wp3 ])' scene='User:Nikos_Pinotsis/Workbench/Obscurin/2wp3/2'> </scene> | ||
| - | [[User:Andrea Ghisleni|Andrea Ghisleni]] 17:09, 23 November 2012 (IST) | ||
===='''Cardiac obscurin schematic structure'''==== | ===='''Cardiac obscurin schematic structure'''==== | ||
[[Image:obscurindomains.jpg|888px x 290px| ]] | [[Image:obscurindomains.jpg|888px x 290px| ]] | ||
Schematic representation of cardiac obscurin domains in different isoforms according to Fukuzawa et al., 2005 <ref>PMID: 16625312</ref> | Schematic representation of cardiac obscurin domains in different isoforms according to Fukuzawa et al., 2005 <ref>PMID: 16625312</ref> | ||
| + | |||
== '''Protein structure''' == | == '''Protein structure''' == | ||
Obscurin is a ~800kDa protein expressed in skeletal and cardiac muscle. Its modular structure resembles the architecture of titin, indeed the NH2-terminal half of the protein consists of 49 Ig-like domains (with the characteristic β-sandwich shape) and 2 Fn3 domains, each one with a length of 88-92 amino acids residues and no linker sequences between subsequent domains (except between Ob2-Ob3 and Ob24-Ob25). Two clusters of these Ig domains show 70-90% homology at the protein level; these are Ob36-42 and Ob9-18. As in other sarcomeric proteins, the Ig domains present a highly stable structural scaffold that is necessary for mechanical stability as well as versatile surface that fits well with the role of binding site for other proteins <ref>PMID: 19466753</ref>. The COOH-terminus shows a less ordered architecture with a non-modular 417 amino acid sequence with several consensus phosphorylation motifs for ERK-kinase (SPXR) <ref>PMID: 11448995</ref>. A PH domain is located upstream of the last tandem Ig domains Ob56-57, preceding the DH/RhoGEF domain. After the Ob51 Ig domain a non-modular structure includes an IQ domain, so called for the conserved Ile-Gln that is part of a well-characterized binding motif for calmodulin or calmodulin-like protein like myosin light chains. In the isoform obscurin B, an additional COOH-terminal part includes two more Ser/Thr Kinase domains, 2 Ig like domains and an Fn3 domain <ref>PMID: 16625312</ref>. This isoform does not include the extremely COOH-terminal non-modular structure that characterizes the isoform A and contains the Ank1.5 binding site. | Obscurin is a ~800kDa protein expressed in skeletal and cardiac muscle. Its modular structure resembles the architecture of titin, indeed the NH2-terminal half of the protein consists of 49 Ig-like domains (with the characteristic β-sandwich shape) and 2 Fn3 domains, each one with a length of 88-92 amino acids residues and no linker sequences between subsequent domains (except between Ob2-Ob3 and Ob24-Ob25). Two clusters of these Ig domains show 70-90% homology at the protein level; these are Ob36-42 and Ob9-18. As in other sarcomeric proteins, the Ig domains present a highly stable structural scaffold that is necessary for mechanical stability as well as versatile surface that fits well with the role of binding site for other proteins <ref>PMID: 19466753</ref>. The COOH-terminus shows a less ordered architecture with a non-modular 417 amino acid sequence with several consensus phosphorylation motifs for ERK-kinase (SPXR) <ref>PMID: 11448995</ref>. A PH domain is located upstream of the last tandem Ig domains Ob56-57, preceding the DH/RhoGEF domain. After the Ob51 Ig domain a non-modular structure includes an IQ domain, so called for the conserved Ile-Gln that is part of a well-characterized binding motif for calmodulin or calmodulin-like protein like myosin light chains. In the isoform obscurin B, an additional COOH-terminal part includes two more Ser/Thr Kinase domains, 2 Ig like domains and an Fn3 domain <ref>PMID: 16625312</ref>. This isoform does not include the extremely COOH-terminal non-modular structure that characterizes the isoform A and contains the Ank1.5 binding site. | ||
| + | |||
| + | |||
| + | <Structure load='2wp3' size='300' side='right' caption='Crystal Structure of the titin Ig-like domain Complex between the titin M10 (blue) - Obscurin like 1 (Red) (PDB entry: [http://www.pdb.org/pdb/explore/explore.do?structureId=2wp3 2wp3 ])' scene='User:Nikos_Pinotsis/Workbench/Obscurin/2wp3/2'> </scene> | ||
| + | |||
| + | |||
==== '''PDB entries''' ==== | ==== '''PDB entries''' ==== | ||
[[1v1c]] - Solution structure os SH3 domain of obscurin, residues 5601-5668 | [[1v1c]] - Solution structure os SH3 domain of obscurin, residues 5601-5668 | ||
Revision as of 16:09, 11 February 2013
Contents |
Obscurin
Introduction
Obscurin is a giant protein expressed in striated muscle cells. It`s one of the three giant scaffold proteins of the striated muscles (the others are titin and nebulin [1] the encoding gene is localized on chromosome 1 (1q42) and the protein product is about 800 kDa with multiple splice variants of different sizes. The protein architecture resembles the modular organization of titin and is composed of multiple Ig-like domains and Fibronectin-like domains arranged in tandem, with most of them encoded on single exons [2]. The obscurin gene (OBSCN) encodes for at least two splice variants, obscurin A and obscurin B[3] the common part of the two variants is composed of up to 67 Ig-like and fibronectin type III domains (Fn3). The COOH-terminal of the protein contains a calmodulin-binding IQ motif, an SH3 domain and a Rho guanine nucleotide exchange factor (GEF) domain followed by a non-modular sequence with multiple phosphorylation sites at the COOH-terminus. In the obscurin B variant, the skipping of exons 90 and 91 links the Ob67 domain to kinase exons leading to the formation of longer variants with two additional Ser/Thr kinase domains which can also be expressed separately from an internal promoter [4] [5]. As scaffold protein of the sarcomere obscurin has several binding partners, mechanical and signaling functions. Obscurin seems to be involved in the correct organization of myofibrils localized in different regions of the sarcomere at different stages of myofibrillogenesis, starting with the subsarcolemmal sarcomeres in early developing hearts [6]. In developing muscles, obscurin is predominantly found at the Z-disk. Later in development, several lines of evidence showed the predominant localization of the protein in the M-band of mature striated muscles. Both localizations seem predominantly determined by the interaction with titin [7]. The presence of the GEF domain at the COOH-terminus suggests a role as a linker between sarcomeric proteins and the G-protein regulated pathways in myofibrils. Moreover, an involvement of obscurin in the lateral alignment of myofibrils and the M-band associated Sarcoplasmic Reticulum (SR) has been suggested [8] due to the highly stable complex formed by the N-terminal domains of obscurin, the C-terminal M10 domain of titin and a linker region of myomesin, which drives the correct alignment of the M-band and SR [9] [10]. Immunofluorescence analysis demonstrated a peripheral localization with respect to the myofibrillar proteins, and therefore a transverse orientation of obscurin with respect to the filament axis, which can then form a link between the myofibril and the SR lipid membranes [11] [12].
Sequence annotation
The aminoacid sequences of human obscurin are deposited in the UniProt database (see Uniprot for [1]) and GenBank GenBank (OBSCN obscurin). The sequence provided in Figure 1 is based on the analysis of the human splice variants [13]. Based on these different splice pathways, at least 4 isoforms are predicted: 2 long isoforms, two shorter isoforms in the extremely C-terminal part are possible but currently uncharacterized.
Cardiac obscurin schematic structure
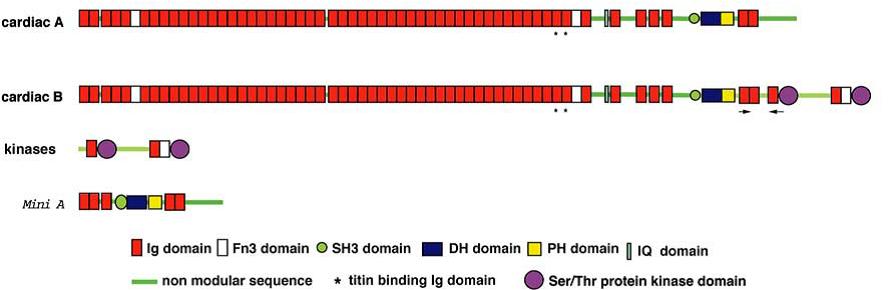 Schematic representation of cardiac obscurin domains in different isoforms according to Fukuzawa et al., 2005 [14]
Schematic representation of cardiac obscurin domains in different isoforms according to Fukuzawa et al., 2005 [14]
Protein structure
Obscurin is a ~800kDa protein expressed in skeletal and cardiac muscle. Its modular structure resembles the architecture of titin, indeed the NH2-terminal half of the protein consists of 49 Ig-like domains (with the characteristic β-sandwich shape) and 2 Fn3 domains, each one with a length of 88-92 amino acids residues and no linker sequences between subsequent domains (except between Ob2-Ob3 and Ob24-Ob25). Two clusters of these Ig domains show 70-90% homology at the protein level; these are Ob36-42 and Ob9-18. As in other sarcomeric proteins, the Ig domains present a highly stable structural scaffold that is necessary for mechanical stability as well as versatile surface that fits well with the role of binding site for other proteins [15]. The COOH-terminus shows a less ordered architecture with a non-modular 417 amino acid sequence with several consensus phosphorylation motifs for ERK-kinase (SPXR) [16]. A PH domain is located upstream of the last tandem Ig domains Ob56-57, preceding the DH/RhoGEF domain. After the Ob51 Ig domain a non-modular structure includes an IQ domain, so called for the conserved Ile-Gln that is part of a well-characterized binding motif for calmodulin or calmodulin-like protein like myosin light chains. In the isoform obscurin B, an additional COOH-terminal part includes two more Ser/Thr Kinase domains, 2 Ig like domains and an Fn3 domain [17]. This isoform does not include the extremely COOH-terminal non-modular structure that characterizes the isoform A and contains the Ank1.5 binding site.
| |||||||||||
