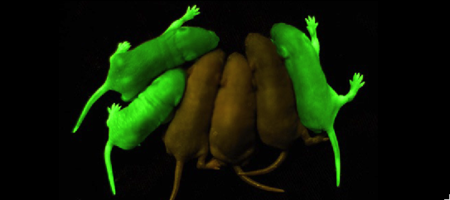
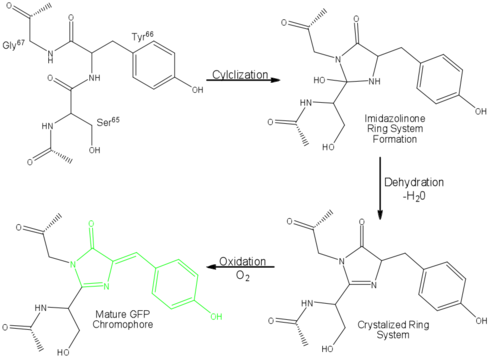
Green Fluorescent Protein
From Proteopedia
| Line 1: | Line 1: | ||
| - | < | + | <StructureSection load='1ema' size='450' side='right' scene='Green_Fluorescent_Protein/1ema_gfp_default/2' caption='Green fluorescent protein complex with peptide-derived chromophore ([[1ema]])' > |
'''Green fluorescent protein (GFP)''' is a [[bioluminescent]] polypeptide consisting of 238 residues isolated from the body of [[Aequorea victoria]] jellyfish.<ref name="PDBsum">[http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1ema&template=main.html], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.</ref> GFP converts the blue chemiluminescent of [[aequorin]] in the jellyfish into green fluorescent light.<ref name="Yang">[http://www-bioc.rice.edu/Bioch/Phillips/Papers/gfpbio.html], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.</ref> It remains unclear why these jellyfish use fluorescence, why green is better than blue, or why they produce a separate protein for green fluorescence as opposed to simply mutating the present aequorin to shift its wavelength,<ref name="Tsien" /> but in the laboratory, GFP can be incorporated into a variety of biological systems in order to function as a marker protein. Since its discovery in 1962, GFP has become a significant contributor to the research of monitoring gene expression, localization, mobility, traffic, interactions between various membrane and cytoplasmic proteins, as well as many others.See also [[Green Fluorescent Protein: Research Tool]].<ref name="Haldar"> [http://www.springerlink.com/content/wvg513864266g77n/fulltext.pdf], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on [http://en.wikipedia.org/wiki/Douglas_Prasher Douglas Prasher], [http://en.wikipedia.org/wiki/Martin_Chalfie Martin Chalfie] and [http://en.wikipedia.org/wiki/Roger_Tsien Roger Tsien].</ref> | '''Green fluorescent protein (GFP)''' is a [[bioluminescent]] polypeptide consisting of 238 residues isolated from the body of [[Aequorea victoria]] jellyfish.<ref name="PDBsum">[http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=1ema&template=main.html], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.</ref> GFP converts the blue chemiluminescent of [[aequorin]] in the jellyfish into green fluorescent light.<ref name="Yang">[http://www-bioc.rice.edu/Bioch/Phillips/Papers/gfpbio.html], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.</ref> It remains unclear why these jellyfish use fluorescence, why green is better than blue, or why they produce a separate protein for green fluorescence as opposed to simply mutating the present aequorin to shift its wavelength,<ref name="Tsien" /> but in the laboratory, GFP can be incorporated into a variety of biological systems in order to function as a marker protein. Since its discovery in 1962, GFP has become a significant contributor to the research of monitoring gene expression, localization, mobility, traffic, interactions between various membrane and cytoplasmic proteins, as well as many others.See also [[Green Fluorescent Protein: Research Tool]].<ref name="Haldar"> [http://www.springerlink.com/content/wvg513864266g77n/fulltext.pdf], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on [http://en.wikipedia.org/wiki/Douglas_Prasher Douglas Prasher], [http://en.wikipedia.org/wiki/Martin_Chalfie Martin Chalfie] and [http://en.wikipedia.org/wiki/Roger_Tsien Roger Tsien].</ref> | ||
| Line 32: | Line 32: | ||
The process is completely auto-catalytic such that there are no known co-factors or enzymatic components required.<ref name="Yang" /> Despite the stability of the final product, while the chromophore is forming, the environmental temperature cannot drop below 30°C or the yield of viable GFP will decrease substantially.<ref name="Yang" /><ref name="Phillips" /> This, of course, is not an issue for the protein in nature as the jellyfish is unlikely to encounter waters of this degree in the Pacific Northwest.<ref name="Tsien" /> Such a temperature sensitivity is only relevant during formation as the stability of the final product is maintained through a network of close contacts surrounding the fluorophore.<ref name="Yang" /> This, however, can and has been used in [[pulse-chase experiments]] in which the GFP-expressing cells are exposed to varying temperatures in place of labeled vs. unlabeled trials.<ref name="Tsien" /> | The process is completely auto-catalytic such that there are no known co-factors or enzymatic components required.<ref name="Yang" /> Despite the stability of the final product, while the chromophore is forming, the environmental temperature cannot drop below 30°C or the yield of viable GFP will decrease substantially.<ref name="Yang" /><ref name="Phillips" /> This, of course, is not an issue for the protein in nature as the jellyfish is unlikely to encounter waters of this degree in the Pacific Northwest.<ref name="Tsien" /> Such a temperature sensitivity is only relevant during formation as the stability of the final product is maintained through a network of close contacts surrounding the fluorophore.<ref name="Yang" /> This, however, can and has been used in [[pulse-chase experiments]] in which the GFP-expressing cells are exposed to varying temperatures in place of labeled vs. unlabeled trials.<ref name="Tsien" /> | ||
| - | |||
| - | <applet load='1ema' size='500' frame='true' align='right' scene='Green_Fluorescent_Protein/Polar_interactions/2' name='2'/> | ||
As the central α-helix is not located directly in the center of the β-barrel, cavities of differing area exist on either side of the chromophore. The larger cavity, consisting of about 135 Å,<ref name="Ormo" /> does not open out to the bulk solvent, but rather houses <scene name='Green_Fluorescent_Protein/Water_molecules/1'>four water molecules</scene>.<ref name="Ormo" /><ref name="Van">van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.</ref> Had this space not been occupied, it would be expected to considerably destabilize the protein as a whole. The hydrogen bonding created by the presence of the water molecules, however, helps to link the buried <scene name='Green_Fluorescent_Protein/Gln69_glu222/1' target='2'>side chains</scene> of Glu<sup>222</sup> and Gln<sup>69</sup> that would otherwise be actively polar.<ref name="Ormo" /> Therefore, the water molecules are extremely important in establishing a hydrogen bonding network about the chromophor.<ref name="Lammich">PMID: 17040991</ref> | As the central α-helix is not located directly in the center of the β-barrel, cavities of differing area exist on either side of the chromophore. The larger cavity, consisting of about 135 Å,<ref name="Ormo" /> does not open out to the bulk solvent, but rather houses <scene name='Green_Fluorescent_Protein/Water_molecules/1'>four water molecules</scene>.<ref name="Ormo" /><ref name="Van">van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.</ref> Had this space not been occupied, it would be expected to considerably destabilize the protein as a whole. The hydrogen bonding created by the presence of the water molecules, however, helps to link the buried <scene name='Green_Fluorescent_Protein/Gln69_glu222/1' target='2'>side chains</scene> of Glu<sup>222</sup> and Gln<sup>69</sup> that would otherwise be actively polar.<ref name="Ormo" /> Therefore, the water molecules are extremely important in establishing a hydrogen bonding network about the chromophor.<ref name="Lammich">PMID: 17040991</ref> | ||
| Line 51: | Line 49: | ||
===Mutant Studies=== | ===Mutant Studies=== | ||
| - | <applet load='1ema' size='400' frame='true' align='right' scene='Green_Fluorescent_Protein/1ema_gfp_barrel/2' name='A'/> | ||
Many mutant green fluorescent proteins have been developed in order to further understand the structure and mechanism of the fluorophore. The first mutagenesis studies simply | Many mutant green fluorescent proteins have been developed in order to further understand the structure and mechanism of the fluorophore. The first mutagenesis studies simply | ||
<scene name='Green_Fluorescent_Protein/Truncated_ends/3' target='A'>truncated the ends</scene> of the amino acid sequence (<scene name='Green_Fluorescent_Protein/1ema_gfp_barrel/2' target='A'>see without truncated ends</scene>. NOTE: The structure represented here is already truncated at the carbonyl terminus). Shortening the polypeptide by more than seven amino acids from either terminus lead to a total loss of fluorescence, as well as a complete failure to absorb light at the traditional wavelengths. This is most likely due to the structure of the protein. The last seven amino acid residues of the carboxyl terminus are roughly disordered, and thus do not interfere with the overall structure. After seven residues, however, the capping α-helix structure is disrupted, leading to an unstable or unformed chromophore. The <scene name='Green_Fluorescent_Protein/Amino_terminus/2' target='A'>amino terminus</scene> is less understood, but the same principle still applies even though the β-barrel does not begin until residue ten or eleven.<ref name="Yang" /> | <scene name='Green_Fluorescent_Protein/Truncated_ends/3' target='A'>truncated the ends</scene> of the amino acid sequence (<scene name='Green_Fluorescent_Protein/1ema_gfp_barrel/2' target='A'>see without truncated ends</scene>. NOTE: The structure represented here is already truncated at the carbonyl terminus). Shortening the polypeptide by more than seven amino acids from either terminus lead to a total loss of fluorescence, as well as a complete failure to absorb light at the traditional wavelengths. This is most likely due to the structure of the protein. The last seven amino acid residues of the carboxyl terminus are roughly disordered, and thus do not interfere with the overall structure. After seven residues, however, the capping α-helix structure is disrupted, leading to an unstable or unformed chromophore. The <scene name='Green_Fluorescent_Protein/Amino_terminus/2' target='A'>amino terminus</scene> is less understood, but the same principle still applies even though the β-barrel does not begin until residue ten or eleven.<ref name="Yang" /> | ||
| Line 67: | Line 64: | ||
A description of some of the ways GFP is being used as a tool in research is at [[Green_Fluorescent_Protein:_Research_Tool]]. | A description of some of the ways GFP is being used as a tool in research is at [[Green_Fluorescent_Protein:_Research_Tool]]. | ||
| - | + | </StructureSection> | |
==3D Structures of Green Fluorescent Protein== | ==3D Structures of Green Fluorescent Protein== | ||
Revision as of 11:39, 20 February 2013
| |||||||||||
Contents |
3D Structures of Green Fluorescent Protein
Update February 2013
2qu1, 2h9w – jGFP - jellyfish
3la1, 3i19, 2wur, 3gex, 2qrf, 2qt2, 2qz0, 2gj1, 2gj2, 3cb9, 3cbe, 3cd1, 3cd9, 2hjo, 2hqz, 2hrs, 2okw, 2oky, 2q57, 2due, 2duf, 2dug, 2duh, 2dui, 2q6p, 2hcg, 2hfc, 2hgd, 2hgy, 2awj, 2awk, 2awl, 2awm, 2g16, 2g2s, 2g3d, 2g5z, 2g6e, 2ah8, 2aha, 2fwq, 2fzu, 2b3p, 2b3q, 1z1p, 1z1q, 1yhg, 1yhh, 1yhi, 1yj2, 1yjf, 1s6z, 1q4a, 1q4b, 1q4c, 1q4d, 1q4e, 1q73, 1qyf, 1qyo, 1qyq, 1qst, 1qy3, 1cv7, 1jc0, 1jc1, 1jby, 1jbz, 1kp5, 1kyp, 1hcj, 1h6r, 1b9c, 1c4f, 1emc, 1eme, 1emf, 1emk, 1eml, 1emm, 2emd, 2emn, 2emo, 1emb, 1gfl, 1ema, 2y0g, 3gj1, 3gj2, 3p28, 1qxt, 3sry, 3ssp, 3st0, 3ufz, 3ug0, 4eul, 4ges – jGFP (mutant)
3evp – jGFP circular permutation
2h6v – jGFP+imidazole derivative
1rm9, 1rmm, 1rmo, 1rmp, 1rrz – jGFP containing fluorotryptophan
2o24, 2o29, 2o2b, 1w7u, 1w7t, 1w7s, 1emg – jGFP (mutant)+imidazole derivative
1kyr – jGFP (mutant)+imidazole derivative+Cu
4gf6 - jGFP (mutant) +Cu
1kys – jGFP (mutant)+imidazole derivative+Zn
3ss0, 3ssh, 3ssk, 3ssl, 3sst, 3ssv, 3ssy, 3sv5, 3svb, 3svc, 3svd, 3sve – jGFP (mutant)+imidazole derivative+ halide
3ogo – jGFP+cGFP nanobody – camel
3g9a, 3k1k – jGFP+minimize nanobody – Lama pacos
2qle – GFP (mutant) – Azotobacter vinelandii
2rh7 – GFP – Renilla reniformis
3adf – monomeric azami green – Galaxea fascicularis
2vzx – GFP DENDRA2 – Dendronephthya
2gw3 – GFP KAEDE – Trachiphyllia geoffroyi
2pox, 2gx0, 2gx2, 2iov, 2ie2 – FP DRONPA – Echinophyllia
2dd7 – CpGFP - Chiridius poppei
2dd9 – CpGFP (mutant)
2c9i – saGFP – sea anemone
1xmz – saGFP (mutant)
2c9j – GFP – Cerianthus membranaceus
2hpw – GFP – Clytia gregaria
2g3o – PpGFP – Pontellina plumata
2g6x, 2g6y – PpGFP (mutant)
3lva, 3lvc, 3lvd – GFP (mutant) – Aequoarea coerulescens
Yellow fluorescent protein
3dpw, 3dpx, 3dpz, 3dq1, 3dq2, 3dq3, 3dq4, 3dq5, 3dq6, 3dq7, 3dq8, 3dq9, 3dqa, 3dqc, 3dqd, 3dqe, 3dqf, 3dqh, 3dqi, 3dqj, 3dqk, 3dql, 3dqm, 3dqn, 3dqo, 3dqu, 1myw, 1huy, 2yfp, 1yfp, 3ed8, 3v3d – jGFP (mutant)
1f09, 1f0b – jGFP (mutant)+imidazole derivative+I
2ogr – Z-FP - Zoanthus
2pxs, 2pxw, 1xa9, 1xae – Z-FP (mutant)
2jad – jGFP/glutaredoxin
Red fluorescent protein
2icr, 2ojk – Z-RFP
2fl1 – Z-RFP (mutant)
3bx9, 3bxa, 3bxb, 3bxc, 3e5t, 3e5w, 1uis, 3ip2, 3pj5, 3pj7, 3pjb, 3pib - EnRFP – Entacmaea quadricolor
3e5v, 3rwt – EnRFP (mutant)
1zgo, 2vad, 2vae, 1ggx – DiRFP – Discosoma
1zgp, 1zgq, 2h8q, 2v4e, 1g7k – DiRFP (mutant)
3cfa – AsRFP – Anemonia sulcata
3nt3, 3nt9 – RFP – artificial gene
1yzw – RFP – Heteractis
3ir8 – RFP – Montipora
4edo, 4eds – GFP – Entacmaea quadricolor
3u8a, 3u8c – GFP - synthetic
Cyan fluorescent protein
2wsn, 2wso, 2ydz - jGFP
2ye0, 2ye1, 3ztf, 4ar7, 4as8 - jGFP (mutant)
2otb – cyan C-FP – Clavularia
2ote - cyan C-FP (mutant)
2zo6, 2zo7 – cyan FP – Fungia concinna
1oxd, 1oxe, 1oxf – cyan FP (mutant) – marker plasmid
Blue fluorescent protein
1bfp – jGFP (mutant)
Photoconvertible fluorescent protein
2vvh, 2vvi, 2vvj, 3p8u – LhGFP (mutant) – Lobophyllia hemprichii
1zux – LhGFP
2btj - LhGFP+imidazole derivative
2ddc, 1xss – FfFP – Favia favus
2ddd - FfFP (mutant)
3cff, 3cfh – AsGFP (mutant)
Green fluorescent protein chimera
3ai4 – jGFP/mPolymerase iota ubiquitin binding motif - mouse
3ai5, 3vht - jGFP/m ubiquitin
3o77, 3o78, 3ek4, 3ek7 - jGFP/myosin light chain kinase/calmodulin
4anj - jGFP/myosin light chain kinase + calmodulin
3evr, 3evu, 3evv - jGFP/myosin light chain kinase/calmodulin+Ca
3ek8, 3ekh, 3ekj - jGFP/myosin light chain kinase/calmodulin (mutant)
3osq, 3osr – jGFP/maltose-binding protein
3u8p – jGFP/cytochrome b562
Reference for this Structure
Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
References
- ↑ 1.0 1.1 [1], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 [2], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 [3], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on Douglas Prasher, Martin Chalfie and Roger Tsien.
- ↑ 5.0 5.1 5.2 5.3 [4], Shimomura O. The discovery of green fluorescent protein. Nobel Prize Lecture; 2009;; biographical background at Wikipedia.
- ↑ [5],Cowles D, Cowles J. Aequorea victoria. 2007. Walla Wall University.
- ↑ Primary structure at www.ebi.aci.uk.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 9.13 Phillips GN Jr. Structure and dynamics of green fluorescent protein. Curr Opin Struct Biol. 1997 Dec;7(6):821-7. PMID:9434902
- ↑ Andrews BT, Gosavi S, Finke JM, Onuchic JN, Jennings PA. The dual-basin landscape in GFP folding. Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12283-8. Epub 2008 Aug 19. PMID:18713871
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 [6],Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien R. 1995. Understanding, improving, and using green fluorescent protein. Trends in Biochemical Sciences. 20(11): 448-455. DOI 0.1016/S0968-0004(00)89099-4.
- ↑ Bublitz G, King BA, Boxer SG. 1998. Electronic structure of the chromophore in green fluorescent protein (GFP). Journal of the American Chemical Society. 120(36): 9370-9371. DOI 10.1021/ja98160e.
- ↑ van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.
- ↑ 14.0 14.1 Lammich L, Petersen MA, Nielsen MB, Andersen LH. The gas-phase absorption spectrum of a neutral GFP model chromophore. Biophys J. 2007 Jan 1;92(1):201-7. Epub 2006 Oct 13. PMID:17040991 doi:10.1529/biophysj.106.093674
- ↑ Information about edge-face (CH/π) interactions.
- ↑ Fang C, Frontiera RR, Tran R, Mathies RA. Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature. 2009 Nov 12;462(7270):200-4. PMID:19907490 doi:10.1038/nature08527
Additional Resources
- For additional information, see: Colored & Bioluminescent Proteins
- First Glance
- PDBsum: 1ema
- RCSB PDB 1ema
- OCA
- UniProt: P42212
- Scop: P42212
- CATH: 1emaA00
- Pfam: PF01353
- InterPro: IPR000786
- GFP featured at the Molecule of the Month series of tutorials by David Goodsell.
- Inside green fluorescent protein - editor's summary that accompanied structural detail of GFP chromophore on the cover of Nature.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Wayne Decatur, Karsten Theis, Eran Hodis, Laura Carbone, Karl Oberholser, Mark Hoelzer, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Marius Mihasan, Joseph M. Steinberger, David Canner