This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2d1r
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:2d1r.png|left|200px]] | ||
| - | |||
{{STRUCTURE_2d1r| PDB=2d1r | SCENE= }} | {{STRUCTURE_2d1r| PDB=2d1r | SCENE= }} | ||
| - | |||
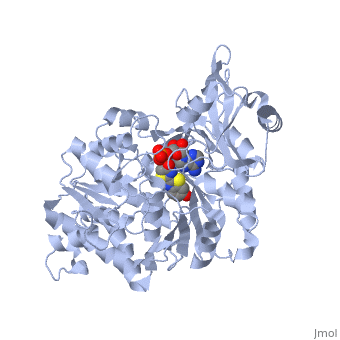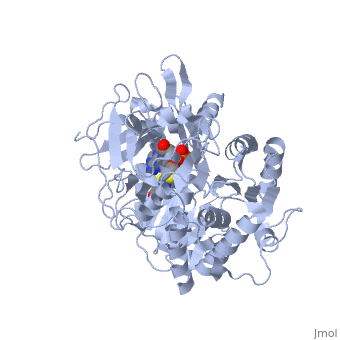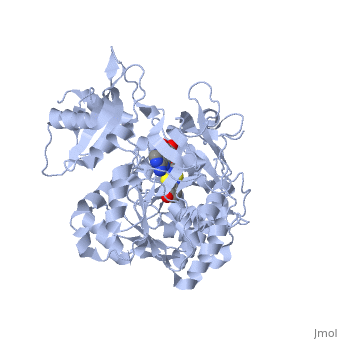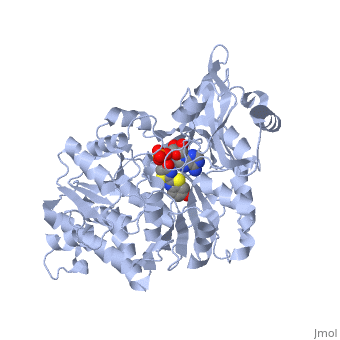
===Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP=== | ===Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP=== | ||
| - | |||
{{ABSTRACT_PUBMED_16541080}} | {{ABSTRACT_PUBMED_16541080}} | ||
Revision as of 10:27, 11 March 2013
| |||||||||
| 2d1r, resolution 1.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Non-Standard Residues: | |||||||||
| Activity: | Photinus-luciferin 4-monooxygenase (ATP-hydrolyzing), with EC number 1.13.12.7 | ||||||||
| Related: | 2d1s, 2d1t | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP
Template:ABSTRACT PUBMED 16541080
About this Structure
2d1r is a 1 chain structure with sequence from Luciola cruciata. Full crystallographic information is available from OCA.
See Also
Reference
- Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006 Mar 16;440(7082):372-6. PMID:16541080 doi:10.1038/nature04542
Categories: Luciola cruciata | Hiratake, J. | Ichiyama, S. | Kato, H. | Kobashi, N. | Nakatsu, T. | RSGI, RIKEN Structural Genomics/Proteomics Initiative. | Sakata, K. | Saldanha, A. | Alpha+beta | Alpha/beta | Beta barrel | Oxidoreductase | Riken structural genomics/proteomics initiative | Rsgi | Structural genomic