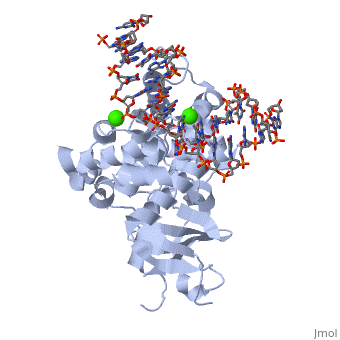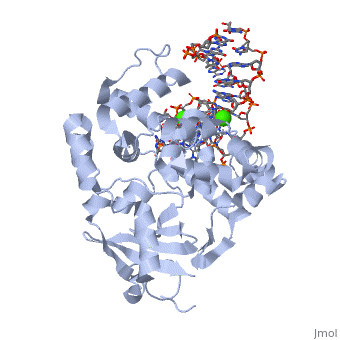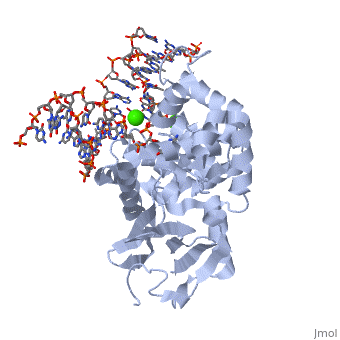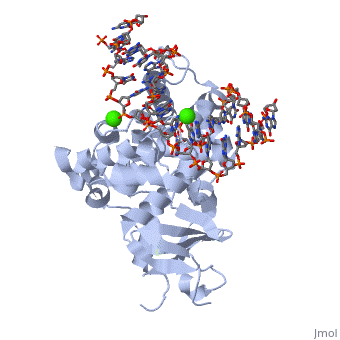
8-oxoguanine glycosylase (hOGG1) is a DNA glycosylase coded from the OGG1 gene in humans; however, many homologs exist in different organisms and this enzyme was originally discovered in yeast[1]. It is responsible for removing genotoxic lesions caused by oxidative damage in the presence of reactive oxygen species (ROS)[1]. Glycosylases, in general, are key enzymes for base excision repair and therefore are essential for maintaining integrity of the genetic material. Once a lesion is successfully excised, the transcription machinery of the cell can repair the DNA strand; however, if it is not repaired mutagenesis occurs possibly leading to cancer and other degenerative diseases[1]. 8-oxoguanine glycosylase complex with DNA and Ca+2 ion (1yqr) is shown.
Function
hOGG1 repairs (8-oxoG, GO); this lesion arises from oxidative attack by ROS on G[1]. It is a particularly dangerous, and stable mutation because GO can Hoogsteen base-pair with adenine causing G:C to T:A tranversions[1]. hOGG1 is able to cleave the N-glycosylic bond between the deoxyribose moiety and GO leaving an apurinic-apyrimdinic (AP) site[1]. It also has the intrinsic ability to cleave the 3’ phosphodiester of the AP site by β-elimination, acting as an AP lyase, and making it a bifunctional glycosylase[1]. hOGG1 has greater affinity for GO when it is complementary to C, and hOGG1 also has catalytic activity towards other lesions such as formamidopyrimidines[1].
Structure
hOGG1, , belongs to a super family of DNA repair enzymes that share a conserved, two-domain fold containing a DNA binding motif followed by a Glycine/Proline rich stretch and a invariant Aspartate[1]. This motif is necessary for interacting with DNA to recognize and catalyze the substrate [2]. As well, OGG1 is associated with two calcium ions that help stabilize the deformed DNA back bone at the site of the extruded lesion[2].
Mechanism
Only 50,000 hOGG1 molecules protect the entire 6,000,000,000 nuclear base-pairs in a diploid cell [3]. For this reason it is obvious that hOGG1 must have an efficient mechanism to catalyze GO and discriminate between GO and G even though they differ by only two atoms at C8 and N7 [4].
Recognition
hOGG1 is able to discriminate GO from G with the help of a single hydrogen bond between a [5]. Additional structural studies have indicated that the GO is extruded from the DNA helix and inserted deeply into a where residues lining the pocket can directly interact to excise the lesion[4].
Catalysis
To excise GO, Lys 249 acts as a nucleophile attacking the C1' carbon in a SN1 reaction, when GO is inserted into the catalytic pocket[5]. Then hOGG1 AP lyase activity uses a conserved lysine residue as a nucleophile to generate a covalently linked enzyme-DNA adduct that undergoes a series of subsequent transformations resulting in DNA strand exscission on the 3’ side of the lesion[5].
Importance of hOGG1
Oxidative mutations in DNA are heavily implicated in aging and cancer so hOGG1 activity is necessary for many organisms to increase longevity. In fact C:G to T:A tranversions, that can be casued by GO, are very common in tumor suppressor genes and human cancers. GO has even been suggested as an indicator for breast cancer where ROS can accumulate[1]. Mutations in glycosylase genes, like hOGG1, have also been linked to increased mutation rates[1].
DNA base damage and repair
(this section was created by Jia Zhou and David Canner)
Oxidative damage, 8-oxo-G and transversion mutations
Reactive oxygen species (ROS), such as hydrogen peroxide, superoxide and hydroxyl radicals, has been linked to chemical carcinogenesis[1]. Unfortunately, ROS are by-products of aerobic respiration and inflammatory responses, and can also be generated by exposure to ionizing radiation and other free radical generating agents; which are inevitable. ROS can escape mitochondria and attack cellular genome, which results in numerous genotoxic adducts and DNA strand breaks[2].
DNA bases are susceptible to ROS-mediated oxidation[3]. The low redox potential of guanine makes this base vulnerable to oxidation and leads to a long list of oxidation products[4], among which 7,8-dihydro-8-oxoguanine (also known as 8-oxoG) is one of the most deleterious and frequently formed products (see Fig 1).
![Fig1 Guanine oxidative products: 8-oxoG and fapyG [3]](/wiki/images/thumb/0/0d/8oxoG_and_fapyG.gif/300px-8oxoG_and_fapyG.gif)
Fig1 Guanine oxidative products: 8-oxoG and fapyG [3]
![Fig2 8-oxoG mispairs to A [3]](/wiki/images/thumb/6/6d/8oxoG-A.gif/300px-8oxoG-A.gif)
Fig2 8-oxoG mispairs to A [3]
The 8-oxoG lesion is particularly deleterious because of its ability to mimic T with its syn conformation, forming a stable 8-oxoG(syn):A(anti) base pair [3](See Fig 2). With this feature, 8-oxoG can efficiently bypass replicative DNA polymerases[5]. Therefore, failure to remove 8-oxoG before replication results in G to T transversion mutations[4](see Fig 3).
GO system, BER and OGG1
Organisms employ the GO system to avoid the highly mutagenic 8-oxoG. The GO system contains MutT, MutM (also known as Fpg) and MutY enzymes in bacteria[7] and the corresponding MTH1, OGG1 and MUTYH (formerly hMYH) in human[8].
MutT/MTH1 hydrolyses 8-oxoGTP and removes it from the nucleotide pool, and thus prevents the incorporation of 8-oxoG into DNA. MutM/OGG1 excises 8-oxoG from the 8-oxoG:C base pairs while MutY/MUTYH removes A from the 8-oxoG:A base pairs in DNA [3]. (see Fig 3).
Base excision repair (BER) is the predominate pathway to deal with DNA oxidation damage.
Simply speaking, BER has four steps: first, glycosylases remove the damaged base and leave an AP site; second, the AP site is cleaved by intrinsic lyase activity of glycosylases or an AP endonuclease, followed by some end cleaning processes; third, a DNA polymerase (e.g. Pol) fills the gas with a new nucleotide; and finally, a ligase seals the nick. The figure shows the first two steps of the pathway[9].(See Fig 4)
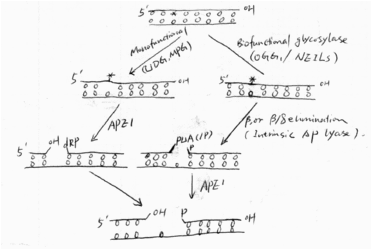
Fig4 First 2 steps in BER Biological function of OGG1
OGG1 (8-oxoG glycosylase) is a glycosylase in base excision repair (BER) pathway that particularly deal with guanine oxidative products. It has been shown that hOGG1 is responsible for most o the glycosylase activity against 8-oxoG in human cells[10]. It removes 8-oxoG from double stranded DNA, and thus prevents the potential G to T transversion mutation.
It was also reported that OGG1 was involved in the expansion of CAG trinucleotide, which is the hallmark of Huntington's disease [11,12]. In addition, OGG1 participates in the regulation of telomere length[13].
OGG1 and carcinogenesis
Considering the high transversion mutation potential of 8-oxoG, people may expect spontaneous carcinogenesis related to OGG1. Surprisingly, no drastic phenotypes were observed in OGG1-/- knockout mice [14]. Although OGG1-/- mice showed a twofold increase in the amount of chromosomal 8-oxoguanine and increased spontaneous mutagenesis, the effect on tumorigenesis was marginal [15,16,17]. However, Sakumi et. al. reported that lung adenoma/carcinoma spontaneously developed in Ogg1 knockout mice approximately 1.5 years after birth, which was five times higher than that observed in wild-type mice[18]. In addition, hOGG1 gene is subject to monoallelic deletion, and nearly 100% of small-cell lung cancers and more than 50% of renal, salivary and non-small-cell cancers lost heterozygosity at this allele[19]. As Bruner et.al. concluded, inactivating mutations in the hOGG1 gene contribute to a small but significant proportion of human cancers[6].
Features and implications of this structure
Overall structure
The α-hOGG1 protein contains 345 amino acids. The (1ebm) was solved by Verdine’s group in 2000. The co-crystal contains the core domain of hOGG1 (residues 12-325, with K249Q substitution) and an oxoG:C-containing duplex 15-base oligomer. It has a resolution of 2.1A.
The protein contains mostly α helices and a few β sheets away from the DNA. The α helices contribute most of the amino acids that interface with the substrate DNA. Interestingly, a large number of the α helices orientate their N-terminal towards the DNA.
The protein latched firmly onto the DNA backbone of the 8-oxoG-containing strand, but not the complementary strand. The 8-oxoG in the substrate duplex is fully extruded from the DNA helix and is inserted to the enzyme active site. The duplex is bended after binding of the enzyme [6].
The structure shows clearly the HhH-GPD (helix-hairpin-helix structure followed by a Gly/Pro-rich loop and a conserved aspartic acid), which is the hallmark of a superfamily of BER proteins typified by endonuclease III and AlkA. The is highlighted in yellow.
Structural implication
Protein-DNA recognition
The oxoG-specificity pocket: The structure clearly shows how cellular repair machinery recognizes oxoG among the vast excess of guanine. Surprisingly, he most characteristic feature of 8-oxoG, the carbonyl group at position 8, does not contribute much to the recognition of 8-oxoG, because the structure shows that carbonyl group is completely devoid of any interacting partner. Instead, the enzyme recognizes the N7-H of 8-oxoG, and form a hydrogen bond to it with the carbonyl group of its Gly 42. On the other hand, the N7 atom of guanine is a proton acceptor instead of proton donor during H-bonding, it cannot form H-bond with Gly 42. Because among all the contacts made to the 8-oxoG, the involvement of Gly 42 by hydrogen bonding is the ONLY clear difference between 8-oxoGuanine and guanine, the role of discrimination of 8-oxoG from G seems to be carried out by a single but critical hydrogen bond[6].
Click to see the 8-oxoG-specific pocket.
Specific recognition of the estranged cytosine
Since the hOGG1 activity on 8-oxoG is highly dependent on the opposite base (with 8-oxoG:C as the best substrate), the recognition of the opposite base might be interesting. This structure also addressed this issue. As Bruner et.al. pointed out, the recognition is achieved by a unique pentad hydrogen bonding system between Cytosine and amino acids from the enzyme: Arg 154 and Arg 204, each forms two H-bond with the opposite Cytosine; and Asn 149 forms one H-bond with the Cytosine. Since only Cytosine has adjacent H-bond acceptor atoms amonge the four DNA bases, the interaction is likely specific. And this explains why the activity of hOGG1 on 8-oxoG depends on the opposite cytosine [6].
Click to see the pentad hydrogen bonds.
The catalytic mechanism
This structure also revealed the components of hOGG1 active site. The proposed nucleophile Lys 249 is calculated to be within reach of the oxoG:C and near the position required for base displacement. Asp 268, which is conserved in HhH-GPD enzymes, is at a suitable position to protonate and deprotonate Lys 249. His270 is positioned reasonably well to protonate O-1’ of the deoxyribose.
Mechanism of hOGG1
Damage Base Search
Based on the structure studies of OGG1 and MutM trapped with DNA, David et.al. proposed an 8-oxo-G lesion search process (shown in fig x)[3]. The 8-oxoG DNA glycosylase moves rapidly along the helix, inserting the probe ligand (for example, Phe 114 in MutM) into the helix to interrogate the base pairs. Intercalation of an amino-acid residue of the enzyme at a normal base pair merely buckles the base pair; however, such a probing event might disrupt an abnormal base pair such as 8-oxoG•C. Such a search process would be extremely fast, so an 8-oxoG base might be missed. However, the research process was proposed to be redundant. When an 8-oxoG encounters hOGG1, it will be expelled from the helix, captured by the ‘exo’ site and passed to the active site. Occasionally expelled undamaged guanine will be captured by ‘exo’ site, but not be processed to the active site; instead, it will be placed back to the helix[3]. (See Fig 5)
![Fig5 A model of damage base searching and recognition [3]](/wiki/images/thumb/2/2c/HOGG1_searching_model.gif/400px-HOGG1_searching_model.gif)
Fig5 A model of damage base searching and recognition [3]
‘Base Flip’
The base extrusion (Base flip) from DNA helix to the enzyme active sites has been described as a ‘pinch-push-plug-pull’ motion. First, glycosylase induces bending and distortion of the DNA double helix (pinch). Second, the glycosylase intercalates an amino acid side chain into the DNA helix, to ‘push’ the target base out of the helix. The same or another intercalating amino acid functions as a ‘plug’ to fill the gap left by the extruded base and stabilizes distorted DNA duplex. A ‘pull’ by active-site residues specific for the relevant target base secures it into the damage base recognition pocket [3,20].
Damage Base recognition
It is a formidable challenge for glycosylase (e.g. hOGG1) to pull out an 8-oxoG base out of a massive amount of normal, undamaged guanine base. Bruner et.al. proposed that the discrimination of 8-oxo-G from guanine might be contributed by a single Hydrogen bonding[6]. While it is possible, this is not likely to be the case. More complex mechanism may be involved. Indeed, feature structural studies revealed more information about damage recognition. For example, using covalent trapping strategy Verdine’s group captured the strucuture of hOGG1 in the act of interrogating normal DNA (G:C)[21]. The structure revealed that even though G is forcibly presented to the enzyme, it does not go into the 8-oxoG specific site but is lodged within an alternative, peripheral ‘exo’ site. Calculations of free energy based on the structure shows a 105-fold preference for 8-oxoG over G in both the active and ‘exo’ sites. Also, the active site of OGG1 has a complementary dipole for 8-oxoG created by a Lys-249-NH3+ and Cys-253-S- amino-acid pair within the active site. And, the contribution of this dipole-dipole interaction to recognition is calculated to be greater than that of the single hydrogen bond to NH7 of 8-oxoG. However, the surrounding structure of Gly42 is very rigid, which presents Gly42 to recognize 8oxoG and reject G from the active site (reviewed by David et.al.[3]).
Click to see hOGG1 with 8-oxoG;
Click to see hOGG1 with Guanine;
Active Site Overview
OGG1 DNA glycosylases have been reported to be structurally related to Endonuclease III and AlkA , all of which containan HhH motif, a glycine/proline-rich loop terminated by an invariant, catalytically essential aspartate residue. It has been noted that a lysine residue within the HhH-GPD motif is conserved in all members of the BER superfamily known to possess efficient lyase activity (Lys249 in hOgg1) and to be absent in all members that lack this activity [22]. Indeed, all biofunctional DNA (glycosylases/ AP lyases activity) of the HhH-GPD family use a lysine residue on the enzyme as the catalytic nucleophile (K249 of hOgg1 or K241 of yeast Ogg1) [22,23,24]. More details about the active site were described in 1.2.
Fig sequence alingment[22]. In addition, His 270 was proposed to act as a proton donor to the oxygen in the deoxyribose ring.
is the geometry of these essential residues.
Enzymatic activity of OGG1
OGG1 majorly catalyzes the cleavage of the guanine oxidative product 8-oxoG and FapyG, with 8-oxoG:C as its best substrate. It is interesting that the enzymatic activity of OGG1 depends essentially on the opposite base of the damaged one. It is also noteworthy that, although OGG1 is a biofuncitonal glycosylase, its lyase activity is very low comparing to its glycosylase activity [25,26,27].
hOGG1 Reaction Mechanism
The mechanism of hOGG1 was proposed to have a similar mechanism as the other biofunctional glycosylases in HhH-GPD superfamily, such as E.coli Fpg [22,28,29]. In the mechanism, an amine nucleophile on the enzyme attacks the glycosidic bond, displacing the damaged base and forming an intermediate in which the enzyme is covalently attached to the deoxyribose moiety of the DNA substrate. Isomerization of the aminal intermediate opens the deoxyribose ring and generates an imine (the Schiff base). The protonated Schiff base act as an electron sink and promotes the abstraction of the 2’-H, which leads to the formation of an enamine. The enamine undergoes conjugate elimination to cleave the 3’C–O bond. Finally, a water molecule comes in and resolve the Shiff base, thus the product is released and the enzyme is recovered [22,28,29,30]. The whole elaborate mechanism derives from the ability of the protonated forms of the Schiff bases to activate the proton abstraction, which initiates the elimination reactions[31]. An amino acid on enzyme may acts as a general acid to protonate the oxygen atom of the deoxyribose ring, and thus facilitates ring open and Shiff Base formation. (Figure 6)
The presence of Schiff base intermediate can be proven by a Schiff base assay. The Shiff base intermediates can be captured when borohydride ( e.g. NaCNBH3) is present in the reaction, since borohydride can reduce the double bond in Schiff bases immediately after they form. The Schiff base is essential for linking glycosylase activity and the lyase activity of a biofunctional glycosylase, and has became a common assay to define a biofunctional glycosylase.
After Nash et.al. cloned hOGG1 in 1996, its catalytic mechanism has been widely researched. Before the resolve of hOGG1 crystal structure, Nash et. al. found that lysine 249 is the critical catalytic amino acid, by using covalent linkage of damage DNA to hOOG1 and Edaman sequencing [22]. This finding well consist with the proposed mechanism of NTH family (hOGG1 was defined as a member of NTH family). And Lysine 249 also seems likely to be the catalytic amino acid that acts as the nucleophile and Shiff base donor in the crystal structure. His270 was proposed to be the amino acid that protonate the oxygen of the deoxyribose ring (See part 2).
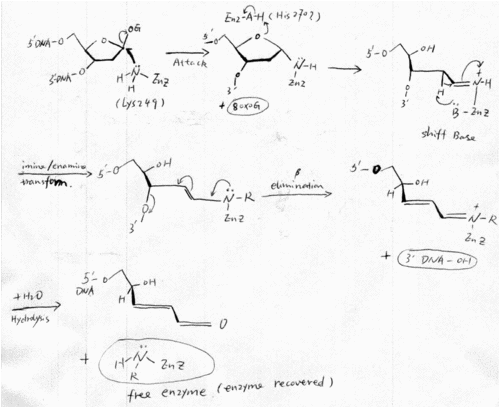
Fig6 Catalytic mechanism of hOGG [22
Similar mechanisms of mOGG1
The amino acid sequence of hOGG1 and mOGG1 are very similar, so are their enzymatic activities and substrate specificities. In addition, the reaction mechanism proposed by Zharkov et.al. is also consistent with that of hOGG1 [27]. In their paper that, three possible pathways were proposed.In all the three cases, the ε-amino group of Lys249 attacks at C1’ of 8-oxodG. A proton donor may interact with O8 (A) or a heterocyclic deoxyribose oxygen (B)to initiate nucleophilic attack at C1'. However, Lys249 may carry out a direct SN2 displacement and form an oxycarbenium intermediate (C). After the attack, 8-oxoG is expelled, and a Schiff base is formed between Lys249 and C19. The Schiff base is hydrolyzed to recover the enzyme after DNA backbone cleavage[27] (see Fig x)
![Three proposed mechanisms of mOGG1 [27]](/wiki/images/thumb/f/fe/MOGG1_mechanism.gif/273px-MOGG1_mechanism.gif)
Three proposed mechanisms of mOGG1 [27]
Monofunctional glycosylase mechanism
The mechanism of monofunctional glycosylase appeared earlier than that of biofunctional glycosylases. The mechanisms of AlkA and UDG are well studied. In the case of these monofunctional glycosylases, the enzymes deliver an activated water molecule to the glycosidic bond of the substrate. The tightly bound water molecule is most likely to perform the [6] nucleophilic attacking to the C1’ of the deoxyribose ring [32].
References
1. Klaunig JE, Kamendulis LM (2004) The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol 44: 239-267.
2. Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709-715.
3. David SS, O'Shea VL, Kundu S (2007) Base-excision repair of oxidative DNA damage. Nature 447: 941-950.
4. Neeley WL, Essigmann JM (2006) Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol 19: 491-505.
5. Scharer OD, Jiricny J (2001) Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays 23: 270-281.
6. Bruner SD, Norman DP, Verdine GL (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403: 859-866.
7. Michaels ML, Miller JH (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol 174: 6321-6325.
8. Barnes DE, Lindahl T (2004) Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet 38: 445-476.
9. Hegde ML, Hazra TK, Mitra S (2008) Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res 18: 27-47.
10. Monden Y, Arai T, Asano M, Ohtsuka E, Aburatani H, et al. (1999) Human MMH (OGG1) type 1a protein is a major enzyme for repair of 8-hydroxyguanine lesions in human cells. Biochem Biophys Res Commun 258: 605-610.
11. Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, et al. (2007) OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447: 447-452.
12. Jarem DA, Wilson NR, Delaney S (2009) Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry 48: 6655-6663.
13. Lu J, Liu Y (2010) Deletion of Ogg1 DNA glycosylase results in telomere base damage and length alteration in yeast. EMBO J 29: 398-409.
14. Tsuzuki T, Nakatsu Y, Nakabeppu Y (2007) Significance of error-avoiding mechanisms for oxidative DNA damage in carcinogenesis. Cancer Sci 98: 465-470.
15. Paz-Elizur T, Sevilya Z, Leitner-Dagan Y, Elinger D, Roisman LC, et al. (2008) DNA repair of oxidative DNA damage in human carcinogenesis: potential application for cancer risk assessment and prevention. Cancer Lett 266: 60-72.
16. Minowa O, Arai T, Hirano M, Monden Y, Nakai S, et al. (2000) Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci U S A 97: 4156-4161.
17. Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, et al. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A 96: 13300-13305.
18. Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, et al. (2003) Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res 63: 902-905.
19. Chevillard S, Radicella JP, Levalois C, Lebeau J, Poupon MF, et al. (1998) Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene 16: 3083-3086.
20. Stivers JT (2004) Site-specific DNA damage recognition by enzyme-induced base flipping. Prog Nucleic Acid Res Mol Biol 77: 37-65.
21. Banerjee A, Yang W, Karplus M, Verdine GL (2005) Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature 434: 612-618.
22. Nash HM, Lu R, Lane WS, Verdine GL (1997) The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: direct identification, ablation and chemical reconstitution. Chem Biol 4: 693-702.
23. van der Kemp PA, Charbonnier JB, Audebert M, Boiteux S (2004) Catalytic and DNA-binding properties of the human Ogg1 DNA N-glycosylase/AP lyase: biochemical exploration of H270, Q315 and F319, three amino acids of the 8-oxoguanine-binding pocket. Nucleic Acids Res 32: 570-578.
24. Guibourt N, Castaing B, Van Der Kemp PA, Boiteux S (2000) Catalytic and DNA binding properties of the ogg1 protein of Saccharomyces cerevisiae: comparison between the wild type and the K241R and K241Q active-site mutant proteins. Biochemistry 39: 1716-1724.
25. Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, et al. (1997) Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J 16: 6314-6322.
26. Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, et al. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol 6: 968-980.
27. Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP (2000) Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J Biol Chem 275: 28607-28617.
28. Dodson ML, Schrock RD, 3rd, Lloyd RS (1993) Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry 32: 8284-8290.
29. Dodson ML, Michaels ML, Lloyd RS (1994) Unified catalytic mechanism for DNA glycosylases. J Biol Chem 269: 32709-32712.
30. Scharer OD, Deng L, Verdine GL (1997) Chemical approaches toward understanding base excision DNA repair. Curr Opin Chem Biol 1: 526-531.
31. Dodson ML, Lloyd RS (2002) Mechanistic comparisons among base excision repair glycosylases. Free Radic Biol Med 32: 678-682.
32. Labahn J, Scharer OD, Long A, Ezaz-Nikpay K, Verdine GL, et al. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell 86: 321-329.
33. Tchou J, Grollman AP (1995) The catalytic mechanism of Fpg protein. Evidence for a Schiff base intermediate and amino terminus localization of the catalytic site. J Biol Chem 270: 11671-11677.
Additional Resources
For additional information, see: Cancer
3D structures of 8-Oxoguanine Glycosylate
DNA glycosylate
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Lu AL, Li X, Gu Y, Wright PM, Chang DY. Repair of oxidative DNA damage: mechanisms and functions. Cell Biochem Biophys. 2001;35(2):141-70. PMID:11892789 doi:10.1385/CBB:35:2:141
- ↑ 2.0 2.1 Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000 Feb 24;403(6772):859-66. PMID:10706276 doi:10.1038/35002510
- ↑ Cappelli E, Hazra T, Hill JW, Slupphaug G, Bogliolo M, Frosina G. Rates of base excision repair are not solely dependent on levels of initiating enzymes. Carcinogenesis. 2001 Mar;22(3):387-93. PMID:11238177
- ↑ 4.0 4.1 Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005 Mar 31;434(7033):612-8. PMID:15800616 doi:10.1038/nature03458
- ↑ 5.0 5.1 5.2 Lingaraju GM, Sartori AA, Kostrewa D, Prota AE, Jiricny J, Winkler FK. A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure. Structure. 2005 Jan;13(1):87-98. PMID:15642264 doi:10.1016/j.str.2004.10.011
- ↑ Dodson ML, Schrock RD 3rd, Lloyd RS. Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry. 1993 Aug 17;32(32):8284-90. PMID:8347626
![Fig1 Guanine oxidative products: 8-oxoG and fapyG [3]](/wiki/images/thumb/0/0d/8oxoG_and_fapyG.gif/300px-8oxoG_and_fapyG.gif)
![Fig2 8-oxoG mispairs to A [3]](/wiki/images/thumb/6/6d/8oxoG-A.gif/300px-8oxoG-A.gif)
![Fig3 human GO system [3]](/wiki/images/thumb/e/e5/Transversion_mutation.gif/376px-Transversion_mutation.gif)

![Fig5 A model of damage base searching and recognition [3]](/wiki/images/thumb/2/2c/HOGG1_searching_model.gif/400px-HOGG1_searching_model.gif)

![Three proposed mechanisms of mOGG1 [27]](/wiki/images/thumb/f/fe/MOGG1_mechanism.gif/273px-MOGG1_mechanism.gif)
