Caspase-3 Regulatory Mechanisms
From Proteopedia
(→Overview of Caspase-3 Structure) |
|||
| Line 24: | Line 24: | ||
== Caspase-3 Loop Bundle and Active Site== | == Caspase-3 Loop Bundle and Active Site== | ||
| - | <StructureSection load=' | + | <StructureSection load='2h5i' size='500' side='right' caption='Structure of Caspase-3 (PDB entry [[2h5i]])' scene='Caspase-3_Regulatory_Mechanisms/Scene1/1'> |
=== Importance of Loop Orientation=== | === Importance of Loop Orientation=== | ||
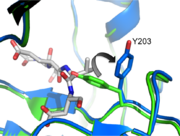
Caspases are extremely dependent on the orientation and geometry of their active site loops. If the loops are not ordered properly the enzyme fails to function. Caspase-3 has four active site loops on each half of the dimer constituting the active site bundle. Proteolytic activity is dependent on cleavage of an intersubunit linker, which releases loop 2 (L2) and L2’. <scene name='Caspase-3_Regulatory_Mechanisms/Scene2_nospin_labels/1'>L2'(green spheres) interacts with the opposite half of the dimer by holding up L2 (blue spheres) </scene>. This allows L2 to make critical contacts with L3 and L4, allowing them to organize the active site, bind substrate, and orient the nucleophilic cysteine 163 (bright green) so that it can cleave after aspartate residues. | Caspases are extremely dependent on the orientation and geometry of their active site loops. If the loops are not ordered properly the enzyme fails to function. Caspase-3 has four active site loops on each half of the dimer constituting the active site bundle. Proteolytic activity is dependent on cleavage of an intersubunit linker, which releases loop 2 (L2) and L2’. <scene name='Caspase-3_Regulatory_Mechanisms/Scene2_nospin_labels/1'>L2'(green spheres) interacts with the opposite half of the dimer by holding up L2 (blue spheres) </scene>. This allows L2 to make critical contacts with L3 and L4, allowing them to organize the active site, bind substrate, and orient the nucleophilic cysteine 163 (bright green) so that it can cleave after aspartate residues. | ||
Revision as of 11:33, 17 March 2013
Contents |
Introduction
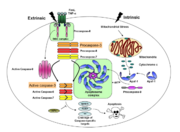
Caspases are cysteine-aspartic acid proteases and are key protein facilitators for the faithful execution of apoptosis or programmed cell death. Dysregulation in the apoptotic pathway has been implicated in a variety of diseases such as neurodegeneration, cancer, heart disease and some metabolic disorders. Because of the crucial role of caspases in the the apoptotic pathway, abnormalities in their functions would cause haywire in the apoptotic cascade and can be deleterious to the cell. Caspases are thus being considered as therapeutic targets in apoptosis-related diseases.
Any apoptotic signal received by the cell causes the activation of initiator caspases (-8 and -9) by associating with other protein platforms to form a functional holoenzyme. These initiator caspases then cleave the executioner caspases -3, -6, and -7. Caspase-3 specifically functions to cleave downstream apoptotic targets as well as both caspase-6 and -7, which in turn cleave their respective targets to induce cell death. Aside from being able to activate caspase-6 and -7, caspase-3 also regulates caspase-9 activity, operating via a feedback loop. This dual action of caspase-3 confers its distinct regulatory mechanisms, resulting in a wider extent of its effects in the apoptotic cascade.
Overview of Caspase-3 Structure
| |||||||||||
Caspase-3 Loop Bundle and Active Site
| |||||||||||
Caspase-3 Regulation
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Scott Eron, Banyuhay P. Serrano, Alexander Berchansky, Yunlong Zhao, Jaime Prilusky, Michal Harel