FK506 binding protein
From Proteopedia
| Line 44: | Line 44: | ||
==Wheat FKBP73 and its comparison with human FKBP52<ref >PMID:20306145</ref>== | ==Wheat FKBP73 and its comparison with human FKBP52<ref >PMID:20306145</ref>== | ||
| - | <StructureSection load='3jym.pdb' size=' | + | <StructureSection load='3jym.pdb' size='350' frame='true' align='right' scene='3jym/Cv/2' caption='Wheat FKBP506 [[3jym]]'> |
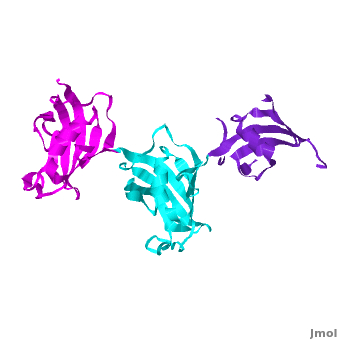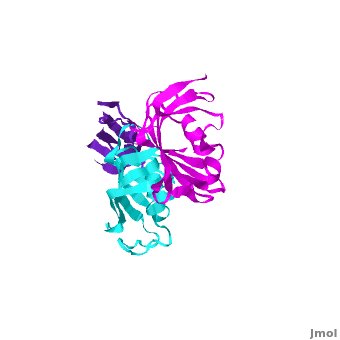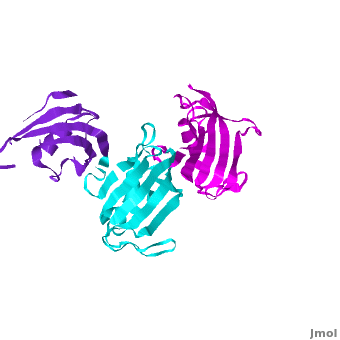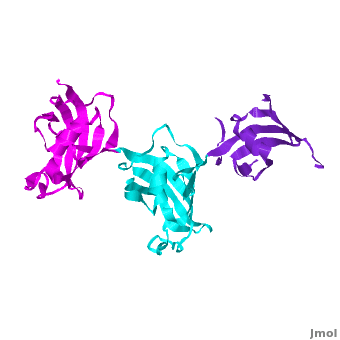
Ribbon representation of the <scene name='3jym/Cv/3'>three FKBP domains</scene>; <font color='blueviolet'><b>wFK73_1 (residues 1–148) in blueviolet</b></font>, <font color='cyan'><b>wFK73_2 (residues 149–266) in cyan</b></font> and <font color='magenta'><b>wFK73_3 (residues 267–386) in magenta</b></font> ([[3jym]]). The wFK73_1 domain exhibits electron density only between residues 33–38, 54–69 and 87–148. The bulges and the flaps as well as the N- and C-termini are labeled. The three FK506 binding (FK) domains of wFKBP73 are held together mainly by <scene name='3jym/Cv/4'>salt bridge networks</scene> situated between each pair of domains. The wFK73_2-wFK73_1 domains are held by a salt bridge between Lys162–Glu62, and a salt bridge network between Arg151–Asp61 and Glu58. The interface between wFK73_2-wFK73_3 is held by two salt bridges between Lys204–Glu269, and Glu178–Lys279. The interactions Lys162–Glu62 and Glu178–Lys279, involve conserved residues (Glu62 from wFK73_1 and Glu178 from wFK73_2, Lys162 from wFK73_2 and Lys279 from wFK73_3). | Ribbon representation of the <scene name='3jym/Cv/3'>three FKBP domains</scene>; <font color='blueviolet'><b>wFK73_1 (residues 1–148) in blueviolet</b></font>, <font color='cyan'><b>wFK73_2 (residues 149–266) in cyan</b></font> and <font color='magenta'><b>wFK73_3 (residues 267–386) in magenta</b></font> ([[3jym]]). The wFK73_1 domain exhibits electron density only between residues 33–38, 54–69 and 87–148. The bulges and the flaps as well as the N- and C-termini are labeled. The three FK506 binding (FK) domains of wFKBP73 are held together mainly by <scene name='3jym/Cv/4'>salt bridge networks</scene> situated between each pair of domains. The wFK73_2-wFK73_1 domains are held by a salt bridge between Lys162–Glu62, and a salt bridge network between Arg151–Asp61 and Glu58. The interface between wFK73_2-wFK73_3 is held by two salt bridges between Lys204–Glu269, and Glu178–Lys279. The interactions Lys162–Glu62 and Glu178–Lys279, involve conserved residues (Glu62 from wFK73_1 and Glu178 from wFK73_2, Lys162 from wFK73_2 and Lys279 from wFK73_3). | ||
| Line 57: | Line 57: | ||
</StructureSection> | </StructureSection> | ||
==SlyD<ref >DOI 10.1007/s00775-011-0855-y</ref>== | ==SlyD<ref >DOI 10.1007/s00775-011-0855-y</ref>== | ||
| - | <StructureSection load='2kr7' size=' | + | <StructureSection load='2kr7' size='350' side='right' scene='Journal:JBIC:14/Cv/2' caption='SlyD [[2kr7]]'> |
SlyD belongs to the FK506-binding protein (FKBP) family with both peptidylprolyl isomerase (PPIase) and chaperone activities, and is considered to be a ubiquitous cytosolic protein-folding facilitator in bacteria. It possesses a histidine- and cysteine-rich C-terminus binding to selected divalent metal ions (''e.g.'', Ni<sup>2+</sup>, Zn<sup>2+</sup>), which is important for its involvement in the maturation processes of metalloenzymes. The solution structure of <scene name='Journal:JBIC:14/Cv/3'>C-terminus-truncated SlyD</scene> from ''Helicobacter pylori'' (HpSlyDΔC) was determined ([[2kr7]]). HpSlyDΔC folds into <scene name='Journal:JBIC:14/Cv/4'>two well-separated, orientation-independent domains:</scene> the <span style="color:cyan;background-color:black;font-weight:bold;">PPIase-active FKBP domain (in cyan)</span> and the <font color='red'><b>chaperone-active insert-in-flap (IF) domain (in red)</b></font>, <font color='darkmagenta'><b>linkers are in darkmagenta</b></font>. The FKBP domain consists of a four-stranded antiparallel <scene name='Journal:JBIC:14/Cv/5'>β-sheet with an α-helix on one side, whereas the IF domain folds into a four-stranded antiparallel β-sheet accompanied by a short α-helix.</scene> Intact ''H. pylori'' SlyD binds both Ni<sup>2+</sup> and Zn<sup>2+</sup>, with dissociation constants of 2.74 and 3.79 μM respectively. Intriguingly, binding of Ni<sup>2+</sup> instead of Zn<sup>2+</sup> induces protein conformational changes around the <scene name='Journal:JBIC:14/Cv/6'>active sites of the FKBP domain, implicating a regulatory role of nickel</scene> <font color='blueviolet'><b>(residues experiencing relatively large chemical shift perturbations upon interactions of HpSlyDΔC with Ni<sup>2+</sup> are in blueviolet)</b></font>. <scene name='Journal:JBIC:14/Cv/7'>The twin-arginine translocation (Tat) signal peptide from the small subunit of [NiFe] hydrogenase (HydA) binds the protein at the IF domain</scene> <font color='orange'><b>(residues in orange)</b></font>. Surprisingly, several residues (Ile41, Gly42, Ile46, and Asn31) were from the FKBP domain, which is likely due to the binding of the longer n-region of HydA Tat peptide to the FKBP domain. Nickel binding and the recognition of the Tat signal peptide by the protein suggest that SlyD participates in [NiFe] hydrogenase maturation processes. | SlyD belongs to the FK506-binding protein (FKBP) family with both peptidylprolyl isomerase (PPIase) and chaperone activities, and is considered to be a ubiquitous cytosolic protein-folding facilitator in bacteria. It possesses a histidine- and cysteine-rich C-terminus binding to selected divalent metal ions (''e.g.'', Ni<sup>2+</sup>, Zn<sup>2+</sup>), which is important for its involvement in the maturation processes of metalloenzymes. The solution structure of <scene name='Journal:JBIC:14/Cv/3'>C-terminus-truncated SlyD</scene> from ''Helicobacter pylori'' (HpSlyDΔC) was determined ([[2kr7]]). HpSlyDΔC folds into <scene name='Journal:JBIC:14/Cv/4'>two well-separated, orientation-independent domains:</scene> the <span style="color:cyan;background-color:black;font-weight:bold;">PPIase-active FKBP domain (in cyan)</span> and the <font color='red'><b>chaperone-active insert-in-flap (IF) domain (in red)</b></font>, <font color='darkmagenta'><b>linkers are in darkmagenta</b></font>. The FKBP domain consists of a four-stranded antiparallel <scene name='Journal:JBIC:14/Cv/5'>β-sheet with an α-helix on one side, whereas the IF domain folds into a four-stranded antiparallel β-sheet accompanied by a short α-helix.</scene> Intact ''H. pylori'' SlyD binds both Ni<sup>2+</sup> and Zn<sup>2+</sup>, with dissociation constants of 2.74 and 3.79 μM respectively. Intriguingly, binding of Ni<sup>2+</sup> instead of Zn<sup>2+</sup> induces protein conformational changes around the <scene name='Journal:JBIC:14/Cv/6'>active sites of the FKBP domain, implicating a regulatory role of nickel</scene> <font color='blueviolet'><b>(residues experiencing relatively large chemical shift perturbations upon interactions of HpSlyDΔC with Ni<sup>2+</sup> are in blueviolet)</b></font>. <scene name='Journal:JBIC:14/Cv/7'>The twin-arginine translocation (Tat) signal peptide from the small subunit of [NiFe] hydrogenase (HydA) binds the protein at the IF domain</scene> <font color='orange'><b>(residues in orange)</b></font>. Surprisingly, several residues (Ile41, Gly42, Ile46, and Asn31) were from the FKBP domain, which is likely due to the binding of the longer n-region of HydA Tat peptide to the FKBP domain. Nickel binding and the recognition of the Tat signal peptide by the protein suggest that SlyD participates in [NiFe] hydrogenase maturation processes. | ||
Revision as of 10:22, 17 April 2013
FK506 binding protein (FKBP) is a prolyl isomerase related to the cyclophilins. FKBP is a folding chaperone for proteins containing prolines. FKBP12 binds the immunosuppressor tacrolimus (FK506) which is used against organ rejection. For more details see
Contents |
Wheat FKBP73 and its comparison with human FKBP52[1]
| |||||||||||
SlyD[2]
| |||||||||||
3D Structures of FKBP
Updated on 17-April-2013
FKPB3
3kz7 – hFKBP FK506-binding domain + immunosuppressant - human
FKPB4
1q1c – hFKBP
1n1a – hFKBP N terminal
1p5q - hFKBP C terminal
1qz2 – hFKBP + Hsp90 peptide
FKBP5
3o5d, 3o5e, 3o5f – hFKBP
3o5g, 3o5i, 3o5j, 3o5k - hFKBP FK506-binding domain
3o5l, 3o5m, 3o5o, 3o5p, 3p5q - hFKBP FK506-binding domain (mutant)
3o5r - hFKBP FK506-binding domain (mutant) + immunosuppressant
FKBP8
2f2d, 3ey6, 2awg - hFKBP FK506-binding domain
2d9f – hFKBP – NMR
2jwx - hFKBP N terminal - NMR
FKBP12
1eym – hFKBP (mutant)
1fkk – hFKBP
2gaq, 2pnu– hFKBP - NMR
1fkd, 1fkj, 2fke, 1qpf, 1qpl – hFKBP + immunosuppressant
2ppp, 2ppn, 2dg3, 1d6o – hFKBP
1j4h, 1j4i – hFKBP + inhibitor
1b6c – hFKBP + TGF-B superfamily receptor I
3fap – hFKBP + FKBP12-rapamycin associated protein
4fap - hFKBP + FKBP12-rapamycin associated protein + immunosuppressant
1tco - FKBP + Ser/Thr phosphatase B2 + immunosuppressant - bovine
1yat – FKBP + antagonist – yeast
FKBP26
3pr9, 3pra, 3prb, 3prd – FKBP – Methanocaldococcus jannaschii
FKBP59
1rot, 1rou – FKBP N terminal – NMR – rabbit
2kr7 – FKBP SlyD – NMR - Helicobacter pylori
2lgo – FKBP – NMR – Giardia lamblia
FKBP73
3jym - FKBP wheat
- ↑ Unger T, Dym O, Albeck S, Jacobovitch Y, Bernehim R, Marom D, Pisanty O, Breiman A. Crystal structure of the three FK506 binding protein domains of wheat FKBP73: evidence for a unique wFK73_2 domain. J Struct Funct Genomics. 2010 Jun;11(2):113-23. Epub 2010 Mar 20. PMID:20306145 doi:10.1007/s10969-010-9085-8
- ↑ Cheng T, Li H, Xia W, Sun H. Multifaceted SlyD from Helicobacter pylori: implication in [NiFe] hydrogenase maturation. J Biol Inorg Chem. 2011 Nov 2. PMID:22045417 doi:10.1007/s00775-011-0855-y