Tropomyosin
From Proteopedia
| Line 11: | Line 11: | ||
== Tropomyosin's Structure == | == Tropomyosin's Structure == | ||
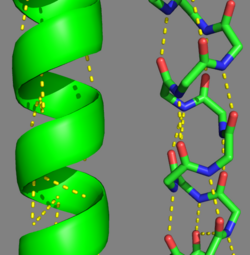
| - | [[Image:Tropomyosin Alpha helix.png | thumb | left | 250x380px | alt text | '''Tropomyosin's''' Alpha Helices are represented in ribbon (Chain A) and ball and stick form (Chain B, without side chains). The yellow lines represent the polar contacts within the protein backbone, which stabilize the helical structure.]]As determined by the '''S'''tructural '''C'''lassification '''o'''f '''P'''rotein's ([http://scop.mrc-lmb.cam.ac.uk/scop/ '''SCOP''']) Database, Tropomyosin is categorized as follows (general to specific): | + | [[Image:Tropomyosin Alpha helix.png | thumb | left | 250x380px | alt text | '''Tropomyosin's''' Alpha Helices are represented in ribbon (Chain A) and ball and stick form (Chain B, without side chains). The yellow lines represent the polar contacts within the protein backbone, which stabilize the helical structure.]] |
| + | {{clear}} | ||
| + | As determined by the '''S'''tructural '''C'''lassification '''o'''f '''P'''rotein's ([http://scop.mrc-lmb.cam.ac.uk/scop/ '''SCOP''']) Database, Tropomyosin is categorized as follows (general to specific): | ||
#'''Class:''' coiled-coil | #'''Class:''' coiled-coil | ||
#'''Fold:''' parallel coiled-coil | #'''Fold:''' parallel coiled-coil | ||
#'''Superfamily:''' tropomyosin | #'''Superfamily:''' tropomyosin | ||
#'''Family:''' pig [[http://www.pdb.org/pdb/explore/explore.do?structureId=1C1G 1c1g]] | #'''Family:''' pig [[http://www.pdb.org/pdb/explore/explore.do?structureId=1C1G 1c1g]] | ||
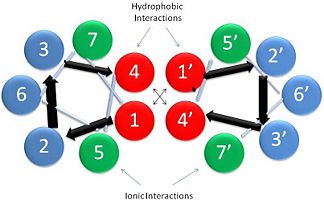
| - | [[Image:Helical Wheel Respresentation of Tropomyosin.jpg | thumb | right | 333x200px | alt text | '''Helical Wheel Diagram Representation of Tropopomyosin:''' The coiled-coil dimer is stabilized by hydrophobic and ionic interactions (red=hydrophobic, blue=polar & green=charged)Image Reconstructed from <ref name="Gunning"/>.]] Categorization of tropomyosin's <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_dimer/2'>coiled-coil</scene> describes a unique pattern of amino acids within the primary structure of the alpha helices that comprise the dimer interface<ref name="Gunning"/>. The unique amino acid pattern, found within all coiled-coil proteins, is a heptad repeat, which follows a similar pattern to: '''H-P-P-H-C-P-C''', where H is hydrophobic, P is polar and C is charged<ref name="Gunning"/><ref name="Whitby"/>. This heptad repeat forms a right-handed alpha helical secondary structure (see right for alpha helix secondary structure)<ref name="Gunning"/>. This alpha helix is special in that it forms a hydrophobic strip along one side, which will interface with an adjacent alpha helix that also contains the heptad repeat and hydrophobic strip. These strips aid in the dimerization of tropomyosin and is important in the characteristic coiled-coil domain. | + | [[Image:Helical Wheel Respresentation of Tropomyosin.jpg | thumb | right | 333x200px | alt text | '''Helical Wheel Diagram Representation of Tropopomyosin:''' The coiled-coil dimer is stabilized by hydrophobic and ionic interactions (red=hydrophobic, blue=polar & green=charged)Image Reconstructed from <ref name="Gunning"/>.]] |
| + | {{clear}} | ||
| + | Categorization of tropomyosin's <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_dimer/2'>coiled-coil</scene> describes a unique pattern of amino acids within the primary structure of the alpha helices that comprise the dimer interface<ref name="Gunning"/>. The unique amino acid pattern, found within all coiled-coil proteins, is a heptad repeat, which follows a similar pattern to: '''H-P-P-H-C-P-C''', where H is hydrophobic, P is polar and C is charged<ref name="Gunning"/><ref name="Whitby"/>. This heptad repeat forms a right-handed alpha helical secondary structure (see right for alpha helix secondary structure)<ref name="Gunning"/>. This alpha helix is special in that it forms a hydrophobic strip along one side, which will interface with an adjacent alpha helix that also contains the heptad repeat and hydrophobic strip. These strips aid in the dimerization of tropomyosin and is important in the characteristic coiled-coil domain. | ||
<br /> | <br /> | ||
This coiled-coil domain is represented as a helical wheel diagram (see right), whereby the four amino acids (two from each alpha chain) that are adjacent to each other contribute to a <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_hydrophobic_aa/4'>hydrophobic interaction</scene>, represented in red, while the four amino acids (two from each alpha chain) flanking the hydrophobic core provide an ionic interaction, or salt-bridge represented as green. The <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_ionic_interaction/2'>salt-bridge</scene> can be observed by observing chain A, residue 26, which is a glutamic acid and forms a salt bridge with an arginine, 305 on Chain B. This weak stabilizing interaction has both and electrostatic interaction, via the ions, and two hydrogen bonds between the residues. This extra stability aids in its left-handed super-coil conformation. The two hydrogen bonds between the residues have an average distance of 2.85Å. Combined the hydrophobic and ionic interactions contribute significantly to the stability of the dimer<ref name="Gunning"/><ref name="Whitby"/>. | This coiled-coil domain is represented as a helical wheel diagram (see right), whereby the four amino acids (two from each alpha chain) that are adjacent to each other contribute to a <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_hydrophobic_aa/4'>hydrophobic interaction</scene>, represented in red, while the four amino acids (two from each alpha chain) flanking the hydrophobic core provide an ionic interaction, or salt-bridge represented as green. The <scene name='User:Gregory_Hoeprich/Sandbox_1/Tropomyosin_ionic_interaction/2'>salt-bridge</scene> can be observed by observing chain A, residue 26, which is a glutamic acid and forms a salt bridge with an arginine, 305 on Chain B. This weak stabilizing interaction has both and electrostatic interaction, via the ions, and two hydrogen bonds between the residues. This extra stability aids in its left-handed super-coil conformation. The two hydrogen bonds between the residues have an average distance of 2.85Å. Combined the hydrophobic and ionic interactions contribute significantly to the stability of the dimer<ref name="Gunning"/><ref name="Whitby"/>. | ||
| Line 39: | Line 43: | ||
In non-muscle systems, as mentioned above, tropomyosin performs a wide variety of functions inside of cells. One of the specific functions tropomyosin performs within cells is recruitment of different myosin motors. Clayton ''et al'', 2010 showed actin decorated with tropomyosin increased myosin-V (cargo transporter) motility as opposed to bare actin<ref name="Clayton">PMID:20705471</ref>. It was also shown that myosin-I (cargo transporter and tension senor) lost its motility function on actin decorated with tropomyosin<ref name="Clayton"/>. Further evidence from a related paper, Stark ''et al'' 2010, shows tropomyosin increases the affinity of myosin-II to actin filaments<ref name="Stark">PMID:20110347</ref>. So actin decorated with tropomyosin appears to spatially regulate motor activity and thus cellular function. | In non-muscle systems, as mentioned above, tropomyosin performs a wide variety of functions inside of cells. One of the specific functions tropomyosin performs within cells is recruitment of different myosin motors. Clayton ''et al'', 2010 showed actin decorated with tropomyosin increased myosin-V (cargo transporter) motility as opposed to bare actin<ref name="Clayton">PMID:20705471</ref>. It was also shown that myosin-I (cargo transporter and tension senor) lost its motility function on actin decorated with tropomyosin<ref name="Clayton"/>. Further evidence from a related paper, Stark ''et al'' 2010, shows tropomyosin increases the affinity of myosin-II to actin filaments<ref name="Stark">PMID:20110347</ref>. So actin decorated with tropomyosin appears to spatially regulate motor activity and thus cellular function. | ||
| - | + | </StructureSection> | |
== 3D structures of Tropomyosin == | == 3D structures of Tropomyosin == | ||
Revision as of 12:48, 1 May 2013
| |||||||||||
3D structures of Tropomyosin
Updated February 2013
3mtu, 3mud – cTPM alpha-1 – chicken
1ic2 - cTPM alpha-1 (mutant)
3u1a, 3u1c – cTPM α-1 N terminal
3u59 – cTPM β N terminal
2w49, 2w4u – cTnnC+cTnnT+cTnnI+cTPM alpha-1+cActin
2z5h – yTPM alpha-1 N-terminal+C-terminal+GNC4 leucine zipper+TnnT – yeast
2z5i - yTPM alpha-1 N-terminal+C-terminal+GNC4 leucine zipper
2efr, 2efs, 2d3e - raTPM alpha-1 C-terminal+GNC4 leucine zipper – rabbit
4a7f, 4a7h, 4a7l - raTPM α-1 + myosin + actin – Cryo EM
1kql - TPM alpha-1 C-terminal+GNC4 leucine zipper - rat
1mv4 - rTPM alpha-1 C-terminal
2g9j - rTPM alpha-1 TM9A+GNC4
2b9c – rTPM mid region
3azd – rTPM N terminal
1c1g – TPM – pig
2tma – TPM - model
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 1.17 1.18 1.19 1.20 1.21 1.22 1.23 1.24 Tropomyosins. I. Gunning, Peter, 1950- II. Series.[DNLM: 1. Tropomyosin. W1 AD559 v.644 2008 / WE 500 T856 2008]
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Frye J, Klenchin VA, Rayment I. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition . Biochemistry. 2010 Jun 15;49(23):4908-20. PMID:20465283 doi:10.1021/bi100349a
- ↑ 3.0 3.1 3.2 Whitby FG, Phillips GN Jr. Crystal structure of tropomyosin at 7 Angstroms resolution. Proteins. 2000 Jan 1;38(1):49-59. PMID:10651038
- ↑ 4.0 4.1 4.2 Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010 Aug 24;20(16):1423-31. Epub 2010 Aug 12. PMID:20705471 doi:10.1016/j.cub.2010.07.026
- ↑ 5.0 5.1 Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010 Mar 15;21(6):989-1000. Epub 2010 Jan 28. PMID:20110347 doi:10.1091/mbc.E09-10-0852
- ↑ 6.0 6.1 Drees B, Brown C, Barrell BG, Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995 Feb;128(3):383-92. PMID:7844152
- ↑ 7.0 7.1 7.2 7.3 7.4 Lehman W, Galinska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2009 May 15;388(4):673-81. Epub 2009 Mar 31. PMID:19341744 doi:10.1016/j.jmb.2009.03.060
- ↑ 8.0 8.1 Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002 Jan;51(1):1-15. PMID:11810692 doi:10.1002/cm.10014
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Gregory Hoeprich, Jaime Prilusky, David Canner, Joel L. Sussman