Sandbox Reserved 687
From Proteopedia
| Line 19: | Line 19: | ||
The structure of HIV-1 [[reverse transcriptase]] is a dimeric protein with a <scene name='Sandbox_Reserved_687/Rt_with_palm_structure_no_i/1'>66-kD polymerase</scene> (p66) domain and a 51-kD RNase H domain (p51). The enzyme has a <scene name='Sandbox_Reserved_687/Hand-like_structure/1'>hand-like</scene> conformation and possesses one active site and three catalytic functions: RNA-dependent DNA polymerization, RNA hydrolysis, and DNA replication. The p66 domain is an RNA-dependent DNA polymerase which binds viral RNA as well as DNA in its single palm-like active site. This active site of the fingers is lined with <scene name='Sandbox_Reserved_687/Hand-like_structure_active_sit/1'>positive charges</scene> (blue) which interact with the negatively charged phosphodiester backbone of the <scene name='Sandbox_Reserved_687/Hand-like_structure_with_dna/1'>bound DNA/RNA</scene>. This diversity of function is brought about by the flexible nature of the hand-like p66 subunit. The p51 subunit is the RNase H domain which degrades the viral RNA after a daughter DNA strand has been synthesized. A structural overview helps us understand the basis of the high mutability of HIV. RNA, which forms the basis of the viral genetic material, is less stable than DNA and is more subject to modification or damage. In addition the reverse transcriptase polymerase domain lacks proofreading exonuclease activity. This lowers the fidelity of the reverse transcription. (Voet et al. 2008, Abbondanzieri et al. 2008) | The structure of HIV-1 [[reverse transcriptase]] is a dimeric protein with a <scene name='Sandbox_Reserved_687/Rt_with_palm_structure_no_i/1'>66-kD polymerase</scene> (p66) domain and a 51-kD RNase H domain (p51). The enzyme has a <scene name='Sandbox_Reserved_687/Hand-like_structure/1'>hand-like</scene> conformation and possesses one active site and three catalytic functions: RNA-dependent DNA polymerization, RNA hydrolysis, and DNA replication. The p66 domain is an RNA-dependent DNA polymerase which binds viral RNA as well as DNA in its single palm-like active site. This active site of the fingers is lined with <scene name='Sandbox_Reserved_687/Hand-like_structure_active_sit/1'>positive charges</scene> (blue) which interact with the negatively charged phosphodiester backbone of the <scene name='Sandbox_Reserved_687/Hand-like_structure_with_dna/1'>bound DNA/RNA</scene>. This diversity of function is brought about by the flexible nature of the hand-like p66 subunit. The p51 subunit is the RNase H domain which degrades the viral RNA after a daughter DNA strand has been synthesized. A structural overview helps us understand the basis of the high mutability of HIV. RNA, which forms the basis of the viral genetic material, is less stable than DNA and is more subject to modification or damage. In addition the reverse transcriptase polymerase domain lacks proofreading exonuclease activity. This lowers the fidelity of the reverse transcription. (Voet et al. 2008, Abbondanzieri et al. 2008) | ||
| - | ==[[Tenofovir]] (TFV)== | + | Because of the high mutability of HIV, resistance develops quickly when the virus is exposed to antiretroviral drugs. Because HIV is not curable at this point, these drugs are therapeutic and only work to reduce viral load which can effectively reduce the patient’s symptoms and boost his/her immune system. Random mutations in the HIV genetic code can allow strains to develop resistance to antiretroviral drugs. Thus, antiretroviral therapy often consists of several different compounds which effect different aspects of HIV infection, reverse transcription, and integration. Combination drugs both reduce viral load, and delay development of resistance. Complera is a drug that combines the antiretroviral activity of three [[reverse transcriptase]] inhibitors (Fig. 1): emtricitabine (ETB), tenofovir (TFV), and rilpivirine (RPV). ETB is a nucleoside RT inhibitor (NRTI), TFV is a nucleotide RT inhibitors (NtRTI), RPV is a second generation nonnucleoside RT inhibitor (NNRTI). |
| + | |||
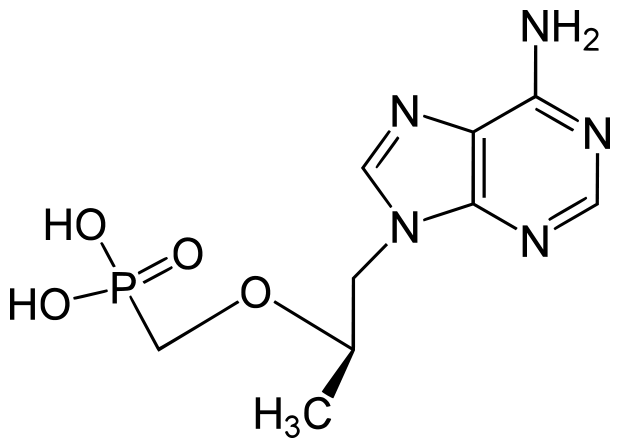
| + | ==[[Tenofovir and Emtricitabine]] (TFV and ETB)== | ||
[[Image:Tenofovir.png]] | [[Image:Tenofovir.png]] | ||
| + | |||
| + | ETB and TFV are highly similar in function and many drug formulations use them in tandem. Once activated by phosphorylation, these compounds compete with naturally occurring dNTPs (Fig. 2) as alternative substrates for viral DNA synthesis by RT. When these compounds are incorporated in the growing chain of viral DNA, they terminate chain extension by functioning as <scene name='Sandbox_Reserved_687/Tenofovir/1'>nucleotide analogues</scene>. In this scene the ligand, TFV, is shown as a space filling molecule in the polymerase site, having stopped the polymerization of additional nucleotides. | ||
| + | |||
| + | <Structure load='ETV' size='300' frame='true' align='right' caption='[[Emtricitabine]] is an analog of cytidine and functions as antiretroviral drug (NRTI) for the treatment of HIV ' scene='Insert optional scene name here' /> | ||
| + | ETB is a cytosine analog and specifically lack a 3’-OH group on its deoxyribose (Singh, 2012). TFV lacks a deoxyribose completely. Both prevent the next nucleotide from binding, as any incoming dNTP cannot form a phosphodiester bond without the presence of a 3’-OH group. Thus, these two compounds act as competitive substrate inhibitors. | ||
| + | |||
| + | |||
==[[Rilpivirine]] (RPV)== | ==[[Rilpivirine]] (RPV)== | ||
| Line 32: | Line 41: | ||
==[[Emtricitabine]] (ECT)== | ==[[Emtricitabine]] (ECT)== | ||
[[Image:Emtricitabine3.png]] | [[Image:Emtricitabine3.png]] | ||
| - | <Structure load='ETV' size='500' frame='true' align='right' caption='[[Emtricitabine]] is an analog of cytidine and functions as antiretroviral drug (NRTI) for the treatment of HIV ' scene='Insert optional scene name here' /> | ||
Revision as of 03:25, 3 May 2013
| This Sandbox is Reserved from 30/01/2013, through 30/12/2013 for use in the course "Biochemistry II" taught by Hannah Tims at the Messiah College. This reservation includes Sandbox Reserved 686 through Sandbox Reserved 700. |
To get started:
More help: Help:Editing |
Contents |
Complera: Emtricitabine/rilpivirine/tenofovir
- Marketed by: Gilead Sciences and Janssen Therapeutics
- Major Indication: HIV infection
- Drug Class: Combination of nucleoside/tide reverse transcriptase inhibitor and nonnucleoside reverse transcriptase inhibitor
- Date of FDA Approval: August 2011
|
Reverse Transcriptase: Upon infection, HIV binds to the CD4 receptor in a helper T cell or other CD4-presenting lymphocyte. The viral envelope is then fused with the cellular membrane allowing intracellular release of viral particles. The utilization of several enzymes helps to solidify the virus’s hold on the cell. HIV reverse transcriptase (RT) works to convert the viral single-stranded RNA (ssRNA) into DNA. HIV integrase then helps to incorporate the newly reverse transcribed DNA into the cellular genome. (Voet et al. 2008)
The structure of HIV-1 reverse transcriptase is a dimeric protein with a (p66) domain and a 51-kD RNase H domain (p51). The enzyme has a conformation and possesses one active site and three catalytic functions: RNA-dependent DNA polymerization, RNA hydrolysis, and DNA replication. The p66 domain is an RNA-dependent DNA polymerase which binds viral RNA as well as DNA in its single palm-like active site. This active site of the fingers is lined with (blue) which interact with the negatively charged phosphodiester backbone of the . This diversity of function is brought about by the flexible nature of the hand-like p66 subunit. The p51 subunit is the RNase H domain which degrades the viral RNA after a daughter DNA strand has been synthesized. A structural overview helps us understand the basis of the high mutability of HIV. RNA, which forms the basis of the viral genetic material, is less stable than DNA and is more subject to modification or damage. In addition the reverse transcriptase polymerase domain lacks proofreading exonuclease activity. This lowers the fidelity of the reverse transcription. (Voet et al. 2008, Abbondanzieri et al. 2008)
Because of the high mutability of HIV, resistance develops quickly when the virus is exposed to antiretroviral drugs. Because HIV is not curable at this point, these drugs are therapeutic and only work to reduce viral load which can effectively reduce the patient’s symptoms and boost his/her immune system. Random mutations in the HIV genetic code can allow strains to develop resistance to antiretroviral drugs. Thus, antiretroviral therapy often consists of several different compounds which effect different aspects of HIV infection, reverse transcription, and integration. Combination drugs both reduce viral load, and delay development of resistance. Complera is a drug that combines the antiretroviral activity of three reverse transcriptase inhibitors (Fig. 1): emtricitabine (ETB), tenofovir (TFV), and rilpivirine (RPV). ETB is a nucleoside RT inhibitor (NRTI), TFV is a nucleotide RT inhibitors (NtRTI), RPV is a second generation nonnucleoside RT inhibitor (NNRTI).
Tenofovir and Emtricitabine (TFV and ETB)
ETB and TFV are highly similar in function and many drug formulations use them in tandem. Once activated by phosphorylation, these compounds compete with naturally occurring dNTPs (Fig. 2) as alternative substrates for viral DNA synthesis by RT. When these compounds are incorporated in the growing chain of viral DNA, they terminate chain extension by functioning as . In this scene the ligand, TFV, is shown as a space filling molecule in the polymerase site, having stopped the polymerization of additional nucleotides.
|
ETB is a cytosine analog and specifically lack a 3’-OH group on its deoxyribose (Singh, 2012). TFV lacks a deoxyribose completely. Both prevent the next nucleotide from binding, as any incoming dNTP cannot form a phosphodiester bond without the presence of a 3’-OH group. Thus, these two compounds act as competitive substrate inhibitors.
Rilpivirine (RPV)
|
RPV in the In red are the binding pocket residues p66 (Leu-100, Lys-101, Lys-103, Val-106, Thr-107, Val-108, Val-179, Tyr-181, Tyr-188, Val-189, Gly-190, Phe-227, Trp-229, Leu- 234, and Tyr-318) and p51(Glu-138). Singh et al.