We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Michael Roberts/BIOL115 Chymo
From Proteopedia
< User:Michael Roberts(Difference between revisions)
| Line 21: | Line 21: | ||
== Beta Barrels, Protein Domains and the Active Center == | == Beta Barrels, Protein Domains and the Active Center == | ||
The chymotrypsin molecule is folded into two <scene name='User:Michael_Roberts/BIOL115_Chymo/2ndry_structure/1'>domains</scene>, each containing six beta strands (orange) arranged as anti-parallel sheets which form a circular structure known as a beta barrel. Rotate the molecule so that you can see down through each of the two beta barrels in turn. | The chymotrypsin molecule is folded into two <scene name='User:Michael_Roberts/BIOL115_Chymo/2ndry_structure/1'>domains</scene>, each containing six beta strands (orange) arranged as anti-parallel sheets which form a circular structure known as a beta barrel. Rotate the molecule so that you can see down through each of the two beta barrels in turn. | ||
| - | |||
| - | The <scene name='User:Michael_Roberts/BIOL115_Chymo/2ndry_structure/2'>active site residues</scene> (Ser-195, His-57 and Asp-102 shown here in spacefill representation), are far apart in the primary sequence but are brought together in a crevice formed between the two beta barrel protein domains. | ||
''Colour key:'' | ''Colour key:'' | ||
{{Template:ColorKey_Helix}}, | {{Template:ColorKey_Helix}}, | ||
{{Template:ColorKey_Strand}}. | {{Template:ColorKey_Strand}}. | ||
| + | |||
| + | The <scene name='User:Michael_Roberts/BIOL115_Chymo/2ndry_structure/2'>active site residues</scene> (Ser-195, His-57 and Asp-102 shown here in spacefill representation), are far apart in the primary sequence but are brought together in a crevice formed between the two beta barrel protein domains. | ||
Current revision
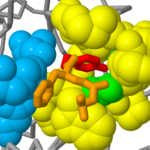
Chymotrypsin active site 1aqf
Chymotrypsin.
Chymotrypsin is a member of a family of enzymes all of which cleave peptide bonds through the action of an active site serine (the serine proteases).
This family includes the pancreatic enzymes chymotrypsin, trypsin and elastase as well as a variety of other proteases (e.g. cocoonase, thrombin, acrosomal protease, etc.). Chymotrypsin, trypsin and elastase show a high degree of similarity in their overall tertiary structure, but have different substrate specificities determined by the different properties of the substrate binding site on each enzyme.
Click on the 'green links' in the text in the scrollable section below to examine this molecule in more detail.
| |||||||||||
