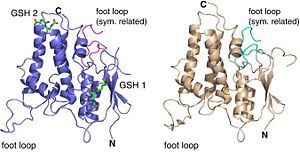
Localization of the different structures of CLIC2
CLIC proteins are a new class of soluble and membrane-bound proteins that have been grouped together on the basis of their sequence similarity. The proteins were named CLIC because the first members of this family to be characterized formed intracellular chloride channels. [1] They display broad tissue and cellular distribution. They have been implicated in kidney function, cell division, and bone resorption. [2] They differ from the other classes of chloride ion channels in primary structure and in the transmembrane regions of the tertiary structure. Since the first member of CLIC, p64 (CLIC5), was discovered in bovine kidney, several members of the CLIC family have been found in other tissues from many species, including NCC27 (CLIC1), CLIC2, CLIC3, mtCLIC (CLIC4), and parchorin (CLIC6). [3] With the exception of p64 and parchorin, these proteins contain a conserved region of approximately 240 residues. [4]
CLIC proteins can localize to distinct cellular membranes, including the nuclear membrane, lysosomal membranes, mitochondria, Golgi membranes, cell–cell junctions, and the plasma membrane. [5]
The CLIC proteins show sequence homology with members of the glutathione-S-transferase (GST) superfamily.
Another feature of CLIC proteins distinguishable from other ion channels is that they exist in two different forms: either as soluble globular proteins, or as an integral membrane protein that is incorporated into lipid bilayers and forms ion channels. [6] These features that are reminiscent of many bacterial pore-forming toxins. [7]
CLIC2 is one of the least characterized CLIC family members. At least two isoforms are known to exist, with the difference being an 18-residue insert occurring immediately after the first β-strand. [8] It has a molecular weight of 28.4 kDa and a calculated isoelectric point of 5.44. [9] Human CLIC2 protein is composed of 247 amino acid residues and is found in many organs, including the spleen, lung, liver, and in both skeletal and cardiac muscles. [10] The CLIC2 gene locates in the telomeric region of Xq28 and is composed of six coding exons and five introns. Since this region of the X chromosome is closely associated with many hereditary diseases, CLIC2 has thus been proposed as a candidate gene for some genetic disorders linked to Xq28.9. [11] Consistent with their high degree of primary structure homology, CLIC2 is similar to CLIC1 and CLIC4 in terms of tertiary structure. Like other members of the CLIC family, CLIC2 can exist as a soluble globular protein, or incorporated into a lipid bilayer to form a Cl– channel. [12]
CLIC2 inhibits cardiac ryanodine receptor (RyR) calciumrelease channels, suggesting that CLIC2 may function to regulate calcium release from intracellular stores in the heart and skeletal muscles. [13] The N-terminal domain of CLIC2 lacks the cysteine equivalent to Cys59 of CLIC1, but contains another cysteine Cys33 that together with the conserved Cys30 (equivalent to Cys24 in CLIC1) forms a CxxC motif similar to glutaredoxin.[14]
Structure
Two different forms of CLIC2 which differ by 18 residues: form A (blue) and form B (grey).
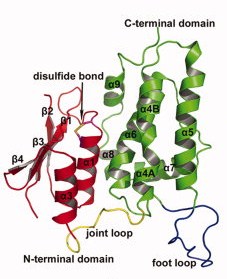
Contrary to each members of the CLICs family, CLIC 2 is a monomer, no matter if it is oxydated or reduiced. It is composed of 247 amino acids, has a weight of 28.4kDa and an isoelectric point at 5.44(crystal structure). The CLIC2 molecule is box shaped (60×60×35 Å) and consists of a four strand core and two helices on one side. Comparing sequence similarities, the core is supposed to adopt the canonical fold of the glutathione S-transferase (GST) superfamily. This has been confirmed by the crystal structure determination of human CLIC1 at 1.4 Å resolution. Then, by analyzing CLIC genes sequences, this protein appears to have two potential transmembrane domains that would correspond to helices α1 and α6 in the GST-like structure of the soluble form. Thanks to immunological, electrophysical and proteolysis studies, we can say that membrane form of CLIC proteins cross the lipid bilayer an odd number of times.
CLIC2 protein is composed of this GST fold and also of two other domains: an N-terminal domain and a C-terminal domain.The (residues 1-94) ) has a thioredoxin-like fold made of four-stranded mixed β-sheets and two α-helices running parallel with the sheet of one face (α1 and α3) and one helix (α2) running perpendicular to the sheet on the other face (β α β α β β).
The (residues 107-245) is exclusively helical composed. It contains a long loop (residues 152-180) between helices 5 and 6, which is a characteristic of the CLIC family wich is called the .
Those two domains are linked by an interdomain loop (residues 95–106) rich in proline residues (more than 33% of proline). Actually there are
, Pro70-Pro71 and Pro96-Pro97 in this loop wich is called . Those diprolines lead to direction changing in the peptide chain. The largest deviations in the N-terminal domain occurs between the residues 55 and 69 that implies helix α2 and its surrounding sequences and also a loop (residues 80-84) that bind the β-strand 4 to helix α3. Crystallographic studies gave two forms of CLIC2, on each one we found out that this protein contain a right handed hook conformation. In fact, the long loop between helices 5 and 6 protrudes on the surface.
Finally, something that we have to highlight is the fact that this protein presents an intramolecular disulfide bridge. It is a well resolved disulfide bond between on the N-terminal of helix 1 thanks to a CXXC motif. The Cys 30 is exposed to the cavity between the N- and the C-terminal domains, which means that this cystein is reachable by other molecules. On the opposite, the Cys 33 is located one turn after the Cys 30, thus is buried inside the N-terminal domain. Let us notice that the partial positive charge present on the N-terminal end of helix increases the nucleophility of the thiol group of the Cys 30, which is a characteristic of protein belonging to the glutaredoxin family. On another hand, we would like to point out the fact that this intramolecular disulfide bridge is the reason why CLIC2 is the only CLIC protein that exists only in the monomer form. In CLIC1, there is no intramolecular disulfide bridge in its monomer state but only in its dimer state where the bound is established between Cys 24 of the first CLIC1 and Cys 59 of the second one. Actually, this bridge is responsible for the dimerisation. In analogy, the corresponding residues in CLIC2 are Cys 30 and Ala 65; consequently, there is no possibility to establish a disulfide bridge. An experiment has been performed to check if CLIC2 cannot dimerise: mutant has been made in which Cys had been settled at the appropriate positions instead of Ala 65. Even in reducing conditions, no disulfide bridge can be created. That means that up to now, CLIC2 remains as a non dimerisable protein.
Actually, the exhibited long loop is flexible and easily affected by crystal packing. When we analyze CLIC2’s electrostatic potential surface, we can observe that contrary to the cavity between the N- and C-terminal domains that is positively charged, the foot loop is negatively charged due to its six acid residues. Indeed, this loop has a crucial functional role; it can be considered as an anchor between CLIC2 and other protein. This foot loop can be inserted into a groove of a neighboring molecule thanks to electrostatic charges. The negative charges provided by the acid resides allow the foot loop to insert into positively charged cavity of another symmetry-related molecule. What’s more, there is an Asp in the 161 position that can form a salt bridge with a Lys from a neighbor molecule, thus the interaction between protein-protein will be increased. Then, the CXXC motif, close to the cavity can provide extensive hydrogen bonds which will stabilize again the interaction. We also can notice that hydrophobic interactions can be involved in the interaction with the highly conserved Ile 158 surrounded by Val 242, Tyr 239 and the alkyl chain of Lys 125 in the groove. This foot loop is an evidence of a structure-function link: the equivalent region in the structurally GST family corresponds to the active site. We can then suggest that CLIC2 will interact with other protein such as the ryanodine receptor for example through the same interaction way.
NB: two crystal forms are obtained differencing by their foot loop position due to limited hydrogen bonding.
Links between CLIC2 and the other CLICs and the GST superfamily members
Similarity to GST Structures
CLIC2 belongs to the glutathione S-transferase (GST) superfamily because it adopts the same secondary structure as GSTs. Between CLIC proteins and GSTs, there are four key residues conserved. A cis-proline residue (Pro 71 in CLIC2) that allows a properly binding site for GSH in all GSTs, an aspartic acid (Asp 183 in CLIC2) that allows interaction between the N-terminal helix and a cap and two glycines that are involved in structural role. The first obvious difference between CLIC2 and other GSTs is, as we said earlier, the fact that CLIC2 is the only one that can exist only as a monomer.
Then, there are two other main differences between GSTs and CLICs. There is a long and highly charged loop region, called the , which we can find only in CLIC2 (from Thr 152 to Thr 180) at the base of the molecule. There is also another charged region in CLIC2 that is absent in other GST proteins. This one is composed by three acid residues in the C-terminal helix (between residues 232 and 239).
On another hand, CLIC2 has a pronounced basic wide crevice like many GST (particularly the beta and the omega classes of GSTs), called the mouth region, which is large enough to accept protein targets. There are some evidences showing that CLIC2 can have an enzymatic activity since this mouth-like region is in the same location as found in GSTs. Actually, the GST active site is composed of a groove between two domains wich forms a GSH binding site (G-site) and a binding site for hydrophobic electrophilic compounds (H-site). However, even with GSH addition in crystallization conditions, no binding of GSH by CLIC2 had been observed. This may be due to the fact that GST residues involved on binding with GSH are nearly all substituted by non conserving residues in CLIC2. Presently, we can only assume that CLIC2 has an enzymatic activity, but no GSH binding site has been discovered yet.
Similarity to other CLIC structures
CLIC4 (2ahe) resolution 1.80Å (residues 58-75)' />
An interesting thing to do is to compare CLIC2 to other CLIC proteins to understand why they are distinguished.
Conserved regions in the glutathione S-transferase (GST) superfamily members appear to have a functional role since CLIC1 can interact with GST thanks to these regions. However, CLIC2 lost most of residues of CLIC1 that interact with GST except Lys 19 ( in CLIC1) and Cys 30 ( in CLIC1). In CLIC1, Lys 13 interacts with a glycyl carboxylate on GSH and Cys 24 forms a mixed disulfide bridge with GSH. The intramolecular disulfide bridge between Cys 30 and Cys 33 in CLIC2 prevents form binding between CLIC2 and GST. As a conclusion, between CLIC1 and CLIC2, the first difference is the loss of the GST binding site in CLIC2.
Up to now, we know the structures of three CLIC proteins, Human CLIC1, Human CLIC2 and Human CLIC4. CLIC2 has 60% sequence identity with CLIC1 and 63% sequence identity with CLIC4. Actually, with both CLIC1 and CLIC4 proteins, most differences are localized in N- and C-terminal regions. The helix α2 with its flanking regions (residues 53-70 corresponding to residues 47-64 in CLIC1 and residues 58-75 in CLIC4) and the foot loop are the most important variable regions.
In CLIC1 and CLIC4 proteins, helix α2 is highly flexible since it can adopt many alternative conformations during crystallization. In CLIC4, this helix seems to be partially disordered.
The foot loop region is also disordered in CLIC4 and is highly mobile too in both CLIC1 and 4 proteins. Although, between CLIC1 and CLIC2, we can notice that there is a short conserved region in the N-terminal end of the foot loop consisting in three residues. We can find the same conformation for Leu 154 (corresponding to Leu 148 in CLIC1) which can lie in a hydrophobic groove between helices α4 and α5. But, the foot loop region of CLIC2 differs from the CLIC1 one because of a diproline, which forms a hairpin structure. In CLIC1, we can find a short helix in the corresponding region. Then, there is another highly conserved residue, Arg171 (Arg 165 in CLIC1) that forms a side chain which interacts with the helix α5.
In CLIC4, we can assume that the foot loop protrudes in a groove belonging to a neighboring molecule too.
Another structural difference between CLIC2 and other CLIC proteins is the fact that helix 4 is a continuous helix while there is a breaking point between residues 112 and 115.
CLIC2 fonctions
It is known that CLIC1 and CLIC4 have a chloride channel role. It has been observed that in liposome, those proteins are responsible for chloride efflux at acid pH. The same has been then demonstrated with CLIC2. Actually, this activity is pH dependant, no activity is observed at pH upper than 8.0.
CLIC2 is a membrane associated protein like another protein called Sedlin. Both proteins seem to interact with each other and have a role in the TRAPP complex or in the targeting and functions of the intracellular chloride channels. TRAPP is a multi sub-unit complex that is involved in intra-cellular communication, in particular to the Golgi apparatus where it always remains bound to the membrane. Let us note that the gene coding for Sedlin is on the X chromosome (like the gene coding for CLIC2), mutations can occur on those genes and lead to a pathology named spondyloepiphyseal dysplasia tarda.
Ryanodine is an alkaloid extracted from plants which modify the activity of intracellular chloride channels like those present on sarcoplasmic reticulum. Those channels are called Ryanodine receptor (RyR). At low concentrations (<10µM), ryanodine opens RyR leading to calcium release in the cytoplasm from the sarcoplasmic reticulum. Whereas at high concentrations (>100µM), ryanodine inhibits RyR. Those receptors are also involved in the physical bound to the sarcoplasmic reticulum and transversal tubules in squeletal muscle cells.
CLIC2 three dimensional fold studies revealed lots of similarities with a number of thioredoxin superfamily members. As it has been described in the first part of the article, CLIC2 N-terminal domain presents similitude with thioltransferase. CLIC2, thanks to its CXXC motif, is assumed to interact with redox proteins. Actually, it has been showed that CLIC2 has a weak peroxidase activity, like many GST family members.
What’s more, CLIC-2 has also a light catalytic activity glutathione transferase.
CLIC2 interacts with the RyR protein (those channels are called Ryanodine receptor RyR). and has by this interaction a huge role in calcium concentration regulation. Actually, CLIC2 is involved in maintaining calcium homeostasy by limiting calcium releases for cellular stock.
Calcium is released from the sarcoplasmic reticulum to the cytoplasm becausee of the opening of RyR channels during the excitation, contraction of cardiac and skeletal muscle cells. CLIC2, by inhibiting cardiac RyR channels in plasmic and sarcoplasmic reticulum membranes, allows stopping this calcium release and getting back to the rest state.
CLIC2 and GST can both inhibits this receptor, that let us suppose that the binding site to RyR is located in a region of the protein with strong similarity. It has been proved that CLIC2 inhibits the cardiac RyR activity in the same way as GST. Experimentaly, adding CLIC2 in the reaction media prevent from excessive release of calcium from the sarcoplasmic reticulum. We also know that the interaction of those proteins does not require oxidation, but is influenced by a conserved Cys residue at position 30.
EM assay and image reconstruction showed that the binding site of CLIC2 on RyR1 is located in an area between domains 5 and 6 in the clamp-shaped region of RyR1.the N-terminal domain and the joint loop of CLIC2 interact with domain 6, where the central region is located, and the foot loop of CLIC2 interacts with domain 5, where the N-terminal region is located.
What’s more, natrin a toxin protein from the venom of the snake Naja naja atra, also binds to domains 5 and 6 of RyR1 concurrently, and inhibits the channel activity of RyR. As a consequence, we can suggest that natrin and CLIC2 have the same action mode using a domain switch mecanism. Domains 9 and 10 in the clamp-shaped region of RyR are thought to be involved in the gating of the RyR channel. Domains 9 and 10 are linked in the close conformation of RyR whereas they are seperated in the open state of RyR, where the RyR channel is open. Previously, it has been shown that the ryanodine binding site on the RyR channel is accessible only when the channel is in an open conformation. When CLIC2 binds to RyR, domains 9 and 10 then get separated. We can say that once CLIC2 is bound on RyR, the RyR channel is in its open conformation although the central channel appears to be closed. This observation is an evidence of the fact that CLIC2 is a RyR channel ligand that does not conform to this general pattern. Usually, the transition of the RyR1 channel from a closed to an open conformation involve specific structural rearrangements : on the one hand there is a modfication in the cytoplasmic clamp region of the channel, and the appearance of a central opening in the transmembrane region o the other hand. Binding of CLIC2 to RyR induces only the separation of domains 9 and 10 but no modification in the transmembrane region. As a consequence, binding of CLIC2 leads to a conformational state of RyR that resembles the open configuration in its clamp-shaped region, which may facilitate the association of ryanodine with the binding site. That can explain how CLIC2 enhances ryanodine effects.
Ryanodine is an alkaloid extracted from plants which modify the activity of intracellular chloride channels like those present on sarcoplasmic reticulum. At low concentrations (<10µM), ryanodine opens RyR leading to calcium release in the cytoplasm from the sarcoplasmic reticulum. Whereas at high concentrations (>100µM), ryanodine inhibits RyR..
Furthermore, the binding of CLIC2 to domains 5 and 6 of RyR increases the interaction between these two domains and so stabilizes the closed state of the RyR channel. This aspect explain how CLIC2 can prevent from Ca2+ efflux from skeletal heavy sarcoplasmic reticulum.
A detail that worth being noticed, it is the fact that a small fraction of cardiac RyR or not totally inhibited by CLIC2. This can be explained by supposing that there exist many isoforms of RyR or simply because we still do not really know if CLIC2 interacts directly with RyR or with other component of the RyR complex.
In addition with CLIC2, ryanodine and GST, RyR has many regulators including Ca2+ and Mg2+, ATP and calmodulin. All these regulators allow RyR channels to respond in synchrony with other cell processes even if CLIC2 remains one of the only a few cytosolic inhibitors of cardiac RyR2 channels, and may suppress their activity during diastole and during stress. The action of CLIC-2 in depressing RyR channel activity and regulating cytoplasmic Ca2+ stores suggests indirectly that it could be effective in preventing or reducing Ca2+ overload in conditions such as ischaemia, and in slowing apoptotic processes.
Those receptors are also involved in the physical bound to the sarcoplasmic reticulum and transversal tubules in squeletal muscle cells
3D structures of Chloride intracellular channel proteins
Ion channels
Additional Resources
For additional information, see: Membrane Channels & Pumps
References
- ↑ Heiss NS, Poustka A. Genomic structure of a novel chloride channel gene, CLIC2, in Xq28. Genomics. 1997 Oct 1;45(1):224-8. PMID:9339381 doi:10.1006/geno.1997.4922
- ↑ Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Board PG, Parker MW. Expression, purification, crystallization and preliminary X-ray diffraction analysis of chloride intracellular channel 2 (CLIC2). Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Nov 1;63(Pt, 11):961-3. Epub 2007 Oct 24. PMID:18007051 doi:10.1107/S1744309107049159
- ↑ Meng X, Wang G, Viero C, Wang Q, Mi W, Su XD, Wagenknecht T, Williams AJ, Liu Z, Yin CC. CLIC2-RyR1 interaction and structural characterization by cryo-electron microscopy. J Mol Biol. 2009 Mar 27;387(2):320-34. Epub 2009 Feb 4. PMID:19356589 doi:10.1016/j.jmb.2009.01.059
- ↑ Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Littler DR, Brown LJ, Mazzanti M, Breit SN, Curmi PM, Dulhunty AF, Board PG, Parker MW. Structure of the Janus protein human CLIC2. J Mol Biol. 2007 Nov 30;374(3):719-31. Epub 2007 Sep 20. PMID:17945253 doi:http://dx.doi.org/10.1016/j.jmb.2007.09.041
- ↑ Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Board PG, Parker MW. Expression, purification, crystallization and preliminary X-ray diffraction analysis of chloride intracellular channel 2 (CLIC2). Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Nov 1;63(Pt, 11):961-3. Epub 2007 Oct 24. PMID:18007051 doi:10.1107/S1744309107049159
- ↑ Meng X, Wang G, Viero C, Wang Q, Mi W, Su XD, Wagenknecht T, Williams AJ, Liu Z, Yin CC. CLIC2-RyR1 interaction and structural characterization by cryo-electron microscopy. J Mol Biol. 2009 Mar 27;387(2):320-34. Epub 2009 Feb 4. PMID:19356589 doi:10.1016/j.jmb.2009.01.059
- ↑ Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Littler DR, Brown LJ, Mazzanti M, Breit SN, Curmi PM, Dulhunty AF, Board PG, Parker MW. Structure of the Janus protein human CLIC2. J Mol Biol. 2007 Nov 30;374(3):719-31. Epub 2007 Sep 20. PMID:17945253 doi:http://dx.doi.org/10.1016/j.jmb.2007.09.041
- ↑ Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Board PG, Parker MW. Expression, purification, crystallization and preliminary X-ray diffraction analysis of chloride intracellular channel 2 (CLIC2). Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Nov 1;63(Pt, 11):961-3. Epub 2007 Oct 24. PMID:18007051 doi:10.1107/S1744309107049159
- ↑ Mi W, Li L, Su XD. 5,5'-Dithio-bis(2-nitrobenzoic acid) modification of cysteine improves the crystal quality of human chloride intracellular channel protein 2. Biochem Biophys Res Commun. 2008 Apr 18;368(4):919-22. Epub 2008 Feb 14. PMID:18280248 doi:10.1016/j.bbrc.2008.02.021
- ↑ Meng X, Wang G, Viero C, Wang Q, Mi W, Su XD, Wagenknecht T, Williams AJ, Liu Z, Yin CC. CLIC2-RyR1 interaction and structural characterization by cryo-electron microscopy. J Mol Biol. 2009 Mar 27;387(2):320-34. Epub 2009 Feb 4. PMID:19356589 doi:10.1016/j.jmb.2009.01.059
- ↑ Mi W, Liang YH, Li L, Su XD. The crystal structure of human chloride intracellular channel protein 2: a disulfide bond with functional implications. Proteins. 2008 Apr;71(1):509-13. PMID:18186468 doi:10.1002/prot.21922
- ↑ Meng X, Wang G, Viero C, Wang Q, Mi W, Su XD, Wagenknecht T, Williams AJ, Liu Z, Yin CC. CLIC2-RyR1 interaction and structural characterization by cryo-electron microscopy. J Mol Biol. 2009 Mar 27;387(2):320-34. Epub 2009 Feb 4. PMID:19356589 doi:10.1016/j.jmb.2009.01.059
- ↑ Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Board PG, Parker MW. Expression, purification, crystallization and preliminary X-ray diffraction analysis of chloride intracellular channel 2 (CLIC2). Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Nov 1;63(Pt, 11):961-3. Epub 2007 Oct 24. PMID:18007051 doi:10.1107/S1744309107049159
- ↑ Cromer BA, Gorman MA, Hansen G, Adams JJ, Coggan M, Littler DR, Brown LJ, Mazzanti M, Breit SN, Curmi PM, Dulhunty AF, Board PG, Parker MW. Structure of the Janus protein human CLIC2. J Mol Biol. 2007 Nov 30;374(3):719-31. Epub 2007 Sep 20. PMID:17945253 doi:http://dx.doi.org/10.1016/j.jmb.2007.09.041
5,5'-Dithio-bis(2-nitrobenzoic acid) modification of cysteine improves the crystal quality of human chloride intracellular channel protein 2.
Mi W, Li L, Su XD.
Biochem Biophys Res Commun. 2008 Apr 18. PMID: 18280248
The crystal structure of human chloride intracellular channel protein 2: a disulfide bond with functional implications.
Mi W, Liang YH, Li L, Su XD.
Proteins. 2008 Apr. PMID: 18186468
Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade
Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, Ardura M, Chung W, Smith E, Wise C, Palucka K, Ramilo O, Punaro M, Banchereau J, Pascual V.
J Exp Med. 2007 Sep 3. PMID: 17724127
A recently identified member of the glutathione transferase structural family modifies cardiac RyR2 substate activity, coupled gating and activation by Ca2+ and ATP
Dulhunty AF, Pouliquin P, Coggan M, Gage PW, Board PG.
Biochem J. 2005 Aug 15.PMID: 15916532
CLIC-2 modulates cardiac ryanodine receptor Ca2+ release channels
Board PG, Coggan M, Watson S, Gage PW, Dulhunty AF.
Int J Biochem Cell Biol. 2004 Aug .PMID: 15147738
Interaction of Sedlin with chloride intracellular channel proteins
Fan L, Yu W, Zhu X.
FEBS Lett. 2003 Apr 10.PMID: 12681486
Differential expression of a chloride intracellular channel gene, CLIC4, in transforming growth factor-beta1-mediated conversion of fibroblasts to myofibroblasts
Rønnov-Jessen L, Villadsen R, Edwards JC, Petersen OW.
Am J Pathol. 2002 Aug .PMID: 12163372
Identification of a novel member of the chloride intracellular channel gene family (CLIC5) that associates with the actin cytoskeleton of placental microvilli
Berryman M, Bretscher A.
Mol Biol Cell. 2000 May.PMID: 10793131