We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Brr2
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='2h9i' size='450' side='right' scene='Sandbox_501/Start_scene/1' caption=''> | ||
='''Structure of Brr2'''= | ='''Structure of Brr2'''= | ||
by Kelly Hrywkiw | by Kelly Hrywkiw | ||
| - | + | ||
__TOC__ | __TOC__ | ||
| Line 13: | Line 14: | ||
=Structure of Brr2= | =Structure of Brr2= | ||
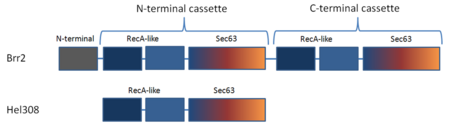
| - | + | [[Image:Brr2 Domain layout 2.PNG|left|450px|thumb|Figure1: Schematic representation of the domain organization in Brr2 and Hel308.]] | |
| - | [[Image:Brr2 Domain layout 2.PNG| | + | |
Brr2 contains an N-terminal domain which is thought to have little tertiary structure (Fig 1)<ref name ="common design"/><ref name ="structural evidence"/>. In addition, is contains two helicase cassettes (residues 496-1310 and 1311-2162) which is outside the norm, as all but one other known DExD/H-box protein contain but one helicase cassette (Fig 2)<ref name ="structural evidence"/>. Each cassette contains dual RecA-like domains and a Sec63 domain (Fig 2)<ref name ="common design"/>. While RecA domains are common to all helicase SF2 proteins Sec63 domain which exhibits a similar sequence to the Sec63 protein, that is essential in the protein-translocation apparatus in the endoplasmic reticulum, are not a common<ref name ="common design"/><ref name ="structural evidence"/>. The RecA-like domain is connected to the Sec63 domain via a WH connector<ref name ="common design"/>. The N-terminal cassette is critical for ATPase activity and U4/U6 unwinding. The C-terminal cassette can undergo catalytically harmful mutations without drastically impairing the overall function of Brr2, and is thought to be involved in protein-protein interactions<ref name ="common design"/><ref name ="structural evidence"/>. | Brr2 contains an N-terminal domain which is thought to have little tertiary structure (Fig 1)<ref name ="common design"/><ref name ="structural evidence"/>. In addition, is contains two helicase cassettes (residues 496-1310 and 1311-2162) which is outside the norm, as all but one other known DExD/H-box protein contain but one helicase cassette (Fig 2)<ref name ="structural evidence"/>. Each cassette contains dual RecA-like domains and a Sec63 domain (Fig 2)<ref name ="common design"/>. While RecA domains are common to all helicase SF2 proteins Sec63 domain which exhibits a similar sequence to the Sec63 protein, that is essential in the protein-translocation apparatus in the endoplasmic reticulum, are not a common<ref name ="common design"/><ref name ="structural evidence"/>. The RecA-like domain is connected to the Sec63 domain via a WH connector<ref name ="common design"/>. The N-terminal cassette is critical for ATPase activity and U4/U6 unwinding. The C-terminal cassette can undergo catalytically harmful mutations without drastically impairing the overall function of Brr2, and is thought to be involved in protein-protein interactions<ref name ="common design"/><ref name ="structural evidence"/>. | ||
==The Sec63 Domain== | ==The Sec63 Domain== | ||
| - | |||
| - | <Structure load='2h9i' size='300' thumb='false' align='left' caption='Figure 2: N terminal (blue) to C terminal (red) ribbon representation of the Sec63 C-terminal domain of Brr2, [[2h9i]]' scene='Sandbox_501/Start_scene/1'/> | ||
| - | |||
To date the only crystal structures of Brr2 are that of the <scene name='Sandbox_501/Start_scene/1'>Sec63</scene> domain in the C-terminal helicase cassette (Sec63c) which contains three sections. The <scene name='Sandbox_501/N_terminus/3'>N-terminal domain</scene>(residues 1859-1990) is comprised of six α helices and one 310 helix. The longest of the helices is α5 which gives stability to the other helices, such that they are able to form a helical bundle through a series of hydrophobic contacts. The <scene name='Sandbox_501/Central_domain/1'>Central domain</scene> (residues 1991-2048) exhibits a helix loop helix fold comprised of four α helices and one 310 helix. The <scene name='Sandbox_501/C_terminus/1'>C-terminal domain</scene> (residues 2049-2163) resembles a seven-stranded immunoglobulin-like β sandwich. Before the N-terminal domain is a region of residues <scene name='Sandbox_501/Flexible_n_domain/1'>(1839-1858)</scene> which is highly flexible and differs between different crystalized versions of Sec63c, however other than this region there is high similarity between the crystal structures. All three domains are in contact with one another. The primary element that appears the fix the domains together is the β sandiwich, specifically the loops connecting β2 and β3, and β6 and β7 which are located towards the center of Sec63c<ref name ="common design"/>. | To date the only crystal structures of Brr2 are that of the <scene name='Sandbox_501/Start_scene/1'>Sec63</scene> domain in the C-terminal helicase cassette (Sec63c) which contains three sections. The <scene name='Sandbox_501/N_terminus/3'>N-terminal domain</scene>(residues 1859-1990) is comprised of six α helices and one 310 helix. The longest of the helices is α5 which gives stability to the other helices, such that they are able to form a helical bundle through a series of hydrophobic contacts. The <scene name='Sandbox_501/Central_domain/1'>Central domain</scene> (residues 1991-2048) exhibits a helix loop helix fold comprised of four α helices and one 310 helix. The <scene name='Sandbox_501/C_terminus/1'>C-terminal domain</scene> (residues 2049-2163) resembles a seven-stranded immunoglobulin-like β sandwich. Before the N-terminal domain is a region of residues <scene name='Sandbox_501/Flexible_n_domain/1'>(1839-1858)</scene> which is highly flexible and differs between different crystalized versions of Sec63c, however other than this region there is high similarity between the crystal structures. All three domains are in contact with one another. The primary element that appears the fix the domains together is the β sandiwich, specifically the loops connecting β2 and β3, and β6 and β7 which are located towards the center of Sec63c<ref name ="common design"/>. | ||
| - | |||
| - | <Structure load='2p6r' size='300' thumb='false' align='right' caption='Figure 2: N terminal (blue) to C terminal (red) ribbon representation of the Sec63 C-terminal domain of Brr2, [[2p6r]]' scene='Sandbox_501/Hel308_start_scene/4'/> | ||
=Structural Similarity to Hel308= | =Structural Similarity to Hel308= | ||
| Line 37: | Line 32: | ||
While the exact sequence and structure of the N-terminal cassette is not a perfect match to Hel308 it is more similar than the C-terminal cassette and therefore may exhibit a similar processive unwinding mechanism that would be important in unwinding the long U4/U6 RNA duplex. The C-terminal cassette has little to no helicase and ATPase activity, however has been shown to interact with other splicing factors such as Prp8 and Snu114<ref name ="structural evidence"/>. This suggest that it may play an important role in protein-protein interactions. | While the exact sequence and structure of the N-terminal cassette is not a perfect match to Hel308 it is more similar than the C-terminal cassette and therefore may exhibit a similar processive unwinding mechanism that would be important in unwinding the long U4/U6 RNA duplex. The C-terminal cassette has little to no helicase and ATPase activity, however has been shown to interact with other splicing factors such as Prp8 and Snu114<ref name ="structural evidence"/>. This suggest that it may play an important role in protein-protein interactions. | ||
| - | + | </StructureSection> | |
=Additional Resources= | =Additional Resources= | ||
*[http://www.rcsb.org/pdb/explore/explore.do?structureId=3HIB Crystal structure of the second Sec63 domain of yeast Brr2, in the RCSB Protein Data Bank] | *[http://www.rcsb.org/pdb/explore/explore.do?structureId=3HIB Crystal structure of the second Sec63 domain of yeast Brr2, in the RCSB Protein Data Bank] | ||
Revision as of 13:21, 10 July 2013
| |||||||||||
Additional Resources
- Crystal structure of the second Sec63 domain of yeast Brr2, in the RCSB Protein Data Bank
- Structure of the C-terminal Sec63 unit of yeast Brr2, P41212 Form, in the RCSB Protein Data Bank
- Structure of the C-terminal Sec63 unit of yeast Brr2, P212121 Form, in the RCSB Protein Data Bank
- DNA REPAIR HELICASE HEL308,in the RCSB Protein Data Bank, in the RCSB Protein Data Bank
- Apo structure of the Hel308 superfamily 2 helicase, in the RCSB Protein Data Bank
- Crystal structure of superfamily 2 helicase Hel308 in complex with unwound DNA, in the RCSB Protein Data Bank
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA. Architecture of the Spliceosome. Biochemistry. 2012 Apr 10. PMID:22471593 doi:10.1021/bi201215r
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Sperling J, Azubel M, Sperling R. Structure and function of the Pre-mRNA splicing machine. Structure. 2008 Nov 12;16(11):1605-15. PMID:19000813 doi:10.1016/j.str.2008.08.011
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009 Jul;16(7):731-9. Epub 2009 Jun 14. PMID:19525970 doi:10.1038/nsmb.1625
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009 Jul;16(7):731-9. Epub 2009 Jun 14. PMID:19525970 doi:10.1038/nsmb.1625
- ↑ 5.0 5.1 5.2 Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007 Jul;14(7):647-52. Epub 2007 Jun 10. PMID:17558417 doi:10.1038/nsmb1246
- ↑ Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, Carter L, Naismith JH, White MF. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008 Feb 22;283(8):5118-26. Epub 2007 Dec 4. PMID:18056710 doi:10.1074/jbc.M707548200