Trypsin
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='13ljj' size='450' side='right' scene='' caption=''> | ||
| + | |||
'''Trypsin''' is a medium size globular protein that functions as a pancreatic serine protease. This enzyme hydrolyzes bonds by cleaving peptides on the C-terminal side of the amino acid residues lysine and arginine. It has also been shown that cleavage will not occur if there is a proline residue on the carboxyl side of the cleavage site. Trypsin was first discovered in 1876 by Kuhne, who investigated the proteolytic activity of the enzyme. In 1931 the enzyme was purified by crystallization by Norothrop and Kunitz and later in 1974 the three dimensional structure of trypsin was determined. Throughout the 1990's the role of trypsin in hereditary pancreatitis and the mutation that causes it was discovered. Today trypsin is used in the development of cell and tissue protocols, as well as in the medical field to determine the role of trypsin in pancreatic diseases<ref>Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html></ref>. | '''Trypsin''' is a medium size globular protein that functions as a pancreatic serine protease. This enzyme hydrolyzes bonds by cleaving peptides on the C-terminal side of the amino acid residues lysine and arginine. It has also been shown that cleavage will not occur if there is a proline residue on the carboxyl side of the cleavage site. Trypsin was first discovered in 1876 by Kuhne, who investigated the proteolytic activity of the enzyme. In 1931 the enzyme was purified by crystallization by Norothrop and Kunitz and later in 1974 the three dimensional structure of trypsin was determined. Throughout the 1990's the role of trypsin in hereditary pancreatitis and the mutation that causes it was discovered. Today trypsin is used in the development of cell and tissue protocols, as well as in the medical field to determine the role of trypsin in pancreatic diseases<ref>Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html></ref>. | ||
{{TOC limit|limit=2}} | {{TOC limit|limit=2}} | ||
| Line 7: | Line 9: | ||
== Ligand Binding and Catalysis == | == Ligand Binding and Catalysis == | ||
| - | + | ||
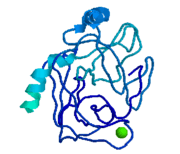
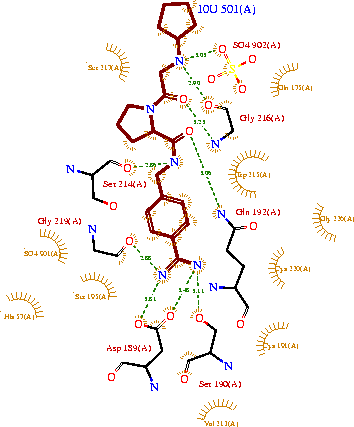
The structure of this particular bovine trypsin was determined in complex with <scene name='Sandbox_45/Btligand/1'>UB-THR 10</scene>, formula '''C'''20'''H'''29'''N'''5'''O'''2, along with two <scene name='Sandbox_45/Btsulfates/1'>sulfate ions</scene>(highlighted) and a Calcium ion (green). Four key amino acids interact with Calcium at a <scene name='Sandbox_45/Lig-metal/1'>subsite loop</scene>. The binding of ligand UB-THR 10 involves <scene name='Sandbox_45/Ligandwaterbridge/1'>water bridges</scene>, direct <scene name='Sandbox_45/Ligandhbond/1'>hydrogen bonding</scene>, and a host of <scene name='Sandbox_45/Ligandhydrophobic/1'>hydrophobic interactions</scene>. The figure below shows this binding in two dimensions. | The structure of this particular bovine trypsin was determined in complex with <scene name='Sandbox_45/Btligand/1'>UB-THR 10</scene>, formula '''C'''20'''H'''29'''N'''5'''O'''2, along with two <scene name='Sandbox_45/Btsulfates/1'>sulfate ions</scene>(highlighted) and a Calcium ion (green). Four key amino acids interact with Calcium at a <scene name='Sandbox_45/Lig-metal/1'>subsite loop</scene>. The binding of ligand UB-THR 10 involves <scene name='Sandbox_45/Ligandwaterbridge/1'>water bridges</scene>, direct <scene name='Sandbox_45/Ligandhbond/1'>hydrogen bonding</scene>, and a host of <scene name='Sandbox_45/Ligandhydrophobic/1'>hydrophobic interactions</scene>. The figure below shows this binding in two dimensions. | ||
| Line 21: | Line 23: | ||
==Catalytic Mechanism== | ==Catalytic Mechanism== | ||
| - | + | [[Image:Serine_protease_mechanism_by_snellios.png |thumb|left|Serine Protease Mechanism]] | |
| - | [[Image:Triad_1.jpg|thumb| | + | [[Image:Triad_1.jpg|thumb|left|Catalytic Triad]] |
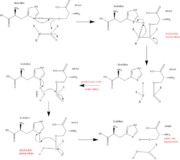
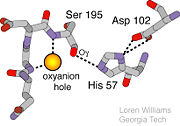
The function of Trypsin is to break down peptides using a hydrolysis reaction into amino acid building blocks. This mechanism is a general catalytic mechanism that all Serine proteases use. The active site where this mechanism occurs in Trypsin is composed of three amino acids and called a catalytic triad. The three catalytic residues are Serine 195, Histidine 57, and Aspartate 102 <ref>Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.</ref>. The structure of the catalytic triad and the mechanism are shown in the figures to the right. In the mechanism, serine is bonded to the imidazole ring of the histidine. When histidine accepts a proton from serine an alkoxide nucleophile is formed. This nucleophile attacks the substrate when the substrate is present. The role of the aspartate residue is hold histidine in the proper position to make it a good proton acceptor. What makes this mechanism works is that a pocket if formed from the three residues and the three residues function to hold each other in proper position for nucleophilic attack. | The function of Trypsin is to break down peptides using a hydrolysis reaction into amino acid building blocks. This mechanism is a general catalytic mechanism that all Serine proteases use. The active site where this mechanism occurs in Trypsin is composed of three amino acids and called a catalytic triad. The three catalytic residues are Serine 195, Histidine 57, and Aspartate 102 <ref>Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.</ref>. The structure of the catalytic triad and the mechanism are shown in the figures to the right. In the mechanism, serine is bonded to the imidazole ring of the histidine. When histidine accepts a proton from serine an alkoxide nucleophile is formed. This nucleophile attacks the substrate when the substrate is present. The role of the aspartate residue is hold histidine in the proper position to make it a good proton acceptor. What makes this mechanism works is that a pocket if formed from the three residues and the three residues function to hold each other in proper position for nucleophilic attack. | ||
The steps of the mechanism involve two tetrahedral intermediates and an Acyl-enzyme intermediate <ref>Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.</ref>. The mechanism can be followed in more detail in the figure on the right <ref>Image From: http://www.bmolchem.wisc.edu/courses/spring503/503-sec1/DRAWINGS/503-3a-2serineprotease.jpg</ref>. | The steps of the mechanism involve two tetrahedral intermediates and an Acyl-enzyme intermediate <ref>Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.</ref>. The mechanism can be followed in more detail in the figure on the right <ref>Image From: http://www.bmolchem.wisc.edu/courses/spring503/503-sec1/DRAWINGS/503-3a-2serineprotease.jpg</ref>. | ||
===Oxyanion Hole=== | ===Oxyanion Hole=== | ||
An important motif that is formed in this reaction is an oxyanion hole. This is also shown in the figure to the right <ref> Williams, Loren. Georgia Tech. http://www2.chemistry.gatech.edu/~1W26/bcourse_information/6521/protein/serine_protease/triad_1/html.</ref>. This oxyanion hole is specifically formed between the amide hydrogen atoms of Serine 195 and Glycine 193. This oxyanion hole stabilizes the tetrahedral intermediate through the distribution of negative charge to the cleaved amide <ref>Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.</ref>. | An important motif that is formed in this reaction is an oxyanion hole. This is also shown in the figure to the right <ref> Williams, Loren. Georgia Tech. http://www2.chemistry.gatech.edu/~1W26/bcourse_information/6521/protein/serine_protease/triad_1/html.</ref>. This oxyanion hole is specifically formed between the amide hydrogen atoms of Serine 195 and Glycine 193. This oxyanion hole stabilizes the tetrahedral intermediate through the distribution of negative charge to the cleaved amide <ref>Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.</ref>. | ||
| - | |||
| - | <applet load="2ptc" size="350" color="white" frame="true" align="right" spinBox="true" | ||
| - | caption="β-Trypsin (magenta) BPT1 (wheat) complex with Ca+2 ion [[2ptc]]" /> | ||
==Trypsin-BPTI complex== | ==Trypsin-BPTI complex== | ||
| + | |||
The trypsin backbone is shown in pink and the trypsin inhibitor, BPTI, in yellow (PDB code [[2ptc]]). The <scene name='Serine_Protease/Active_site/3'>active site</scene> residues [Ser195-His57-Asp102-Ser214] are shown in green, the disulfide bond between residues 14-38 is shown in yellow and the Lys 15 sidechain at the specificity site in pink. | The trypsin backbone is shown in pink and the trypsin inhibitor, BPTI, in yellow (PDB code [[2ptc]]). The <scene name='Serine_Protease/Active_site/3'>active site</scene> residues [Ser195-His57-Asp102-Ser214] are shown in green, the disulfide bond between residues 14-38 is shown in yellow and the Lys 15 sidechain at the specificity site in pink. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Comparison to Chymotrypsin and Elastase== | ==Comparison to Chymotrypsin and Elastase== | ||
<applet scene='Sandbox_32/Chymotrypsin/1' size='275' frame='true' align='true' align='left' caption='Structure of Chymotrypsin and Elastase.'/> | <applet scene='Sandbox_32/Chymotrypsin/1' size='275' frame='true' align='true' align='left' caption='Structure of Chymotrypsin and Elastase.'/> | ||
Trypsin, chymotrypsin, and elastase are all digestive enzymes that are produced in the pancreas and catalyze the hydrolysis of peptide bonds. Each of these enzymes has different specificities in regards to the side chains next to the peptide bond. Chymotrypsin prefers a large hydrophobic residue, trypsin is specific for a positively charged residue, and elastase prefers a small neutral residue. Chymotrypsin, trypsin and elastase are all proteins that contain a catalytic mechanism and hydrolyze peptides using the serine protease mechanism. Chymotrypsin and elastase are both homologs of Trypsin since they are 40% alike in structure and composition <ref> Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008. </ref>. In the <scene name='Sandbox_32/Chymotrypsin/2'>Chymotrypsin</scene> structure shown the alpha helices are blue, the beta sheets are green, and the remainder of the protein is red. In the <scene name='Sandbox_32/Elastase/2'>Elastase</scene> structure shown the alpha helices are in red, the beta sheets are yellow, and the remainder of the protein is orange. | Trypsin, chymotrypsin, and elastase are all digestive enzymes that are produced in the pancreas and catalyze the hydrolysis of peptide bonds. Each of these enzymes has different specificities in regards to the side chains next to the peptide bond. Chymotrypsin prefers a large hydrophobic residue, trypsin is specific for a positively charged residue, and elastase prefers a small neutral residue. Chymotrypsin, trypsin and elastase are all proteins that contain a catalytic mechanism and hydrolyze peptides using the serine protease mechanism. Chymotrypsin and elastase are both homologs of Trypsin since they are 40% alike in structure and composition <ref> Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008. </ref>. In the <scene name='Sandbox_32/Chymotrypsin/2'>Chymotrypsin</scene> structure shown the alpha helices are blue, the beta sheets are green, and the remainder of the protein is red. In the <scene name='Sandbox_32/Elastase/2'>Elastase</scene> structure shown the alpha helices are in red, the beta sheets are yellow, and the remainder of the protein is orange. | ||
| - | |||
| - | |||
{{clear}} | {{clear}} | ||
| - | |||
| - | |||
==3D structures of Trypsin== | ==3D structures of Trypsin== | ||
| Line 141: | Line 121: | ||
[[1xvm]], [[1pq8]], [[1fn8]], [[1fy4]], [[1fy5]], [[1gdn]], [[1gdq]], [[1gdu]] – FoTry + polypeptide<br /> | [[1xvm]], [[1pq8]], [[1fn8]], [[1fy4]], [[1fy5]], [[1gdn]], [[1gdq]], [[1gdu]] – FoTry + polypeptide<br /> | ||
[[2f91]] – Try-hepatopancreas - Crayfish | [[2f91]] – Try-hepatopancreas - Crayfish | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==References== | ==References== | ||
{{Reflist}} | {{Reflist}} | ||
| - | |||
| - | |||
[[he: Trypsin (Hebrew)]] | [[he: Trypsin (Hebrew)]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
[[Category:Trypsin]] | [[Category:Trypsin]] | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 11:55, 16 July 2013
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Eran Hodis, Leah Bowlin, David Canner, Glenn Jones, Ben Hallowell, Karl Oberholser, Jaime Prilusky