Nitrite reductase
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='Cca.pdb' size='500' side='right' scene='Journal:JBIC:16/Cv/2' caption=''> | |
| - | + | ||
'''Nitrite reductase''' (NIR) catalyzes the reduction of NO2 to NO. There are 2 classes of NIR: (1) A heme-containing cytochrome Cd type NIR. This enzyme contains 4 heme groups. Its d-type heme group binds NO<sub>2</sub>. (2) A copper-containing NIR which produces NO<sub>2</sub>. Under anaerobic conditions bacteria rely on the reduction of nitrogen oxide species to obtain energy. NIR is part of the nitrogen cycle used fot this purpose. | '''Nitrite reductase''' (NIR) catalyzes the reduction of NO2 to NO. There are 2 classes of NIR: (1) A heme-containing cytochrome Cd type NIR. This enzyme contains 4 heme groups. Its d-type heme group binds NO<sub>2</sub>. (2) A copper-containing NIR which produces NO<sub>2</sub>. Under anaerobic conditions bacteria rely on the reduction of nitrogen oxide species to obtain energy. NIR is part of the nitrogen cycle used fot this purpose. | ||
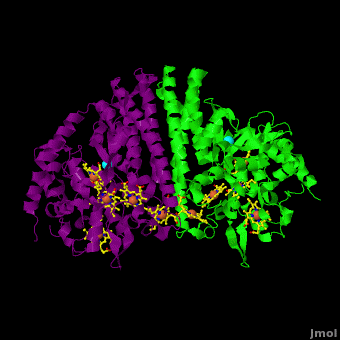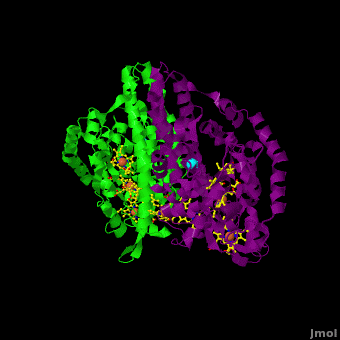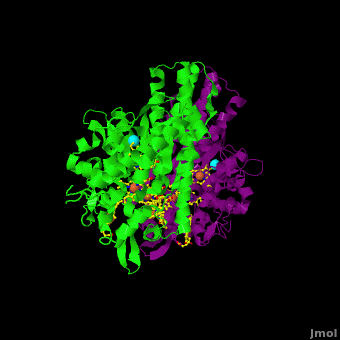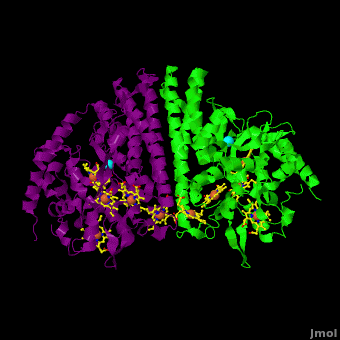
Cytochrome c nitrite reductase (ccNIR) is a central enzyme of the nitrogen cycle. It binds nitrite, and reduces it by transferring 6 electrons to form ammonia. This ammonia can then be utilized to synthesize nitrogen containing molecules such as amino acids or nucleic acids. However, ccNiR’s primary role is to help extract energy from the reduction; ammonia is simply a potentially useful byproduct. In general, heterotrophic organisms feed on electron-rich substances such as sugars or fatty acids. During the metabolism of these substances large numbers of electrons are produced. Many organisms use oxygen as the final acceptor of these electrons, in which case water is formed. However, some organisms can use alternative electron acceptors such as nitrite, which is where ccNiR comes in. | Cytochrome c nitrite reductase (ccNIR) is a central enzyme of the nitrogen cycle. It binds nitrite, and reduces it by transferring 6 electrons to form ammonia. This ammonia can then be utilized to synthesize nitrogen containing molecules such as amino acids or nucleic acids. However, ccNiR’s primary role is to help extract energy from the reduction; ammonia is simply a potentially useful byproduct. In general, heterotrophic organisms feed on electron-rich substances such as sugars or fatty acids. During the metabolism of these substances large numbers of electrons are produced. Many organisms use oxygen as the final acceptor of these electrons, in which case water is formed. However, some organisms can use alternative electron acceptors such as nitrite, which is where ccNiR comes in. | ||
| - | <StructureSection load='Cca.pdb' size='500' side='right' scene='Journal:JBIC:16/Cv/2' caption=''> | ||
'''Laue Crystal Structure of ''Shewanella oneidensis'' Cytochrome c Nitrite Reductase from a High-yield Expression System''' <ref name="Youngblut">doi 10.1007/s00775-012-0885-0</ref> | '''Laue Crystal Structure of ''Shewanella oneidensis'' Cytochrome c Nitrite Reductase from a High-yield Expression System''' <ref name="Youngblut">doi 10.1007/s00775-012-0885-0</ref> | ||
The ccNiR described here is produced by the ''Shewanella oneidensis'' bacterium, which is remarkable in its own right due to the large number of electron acceptors that it can utilize. ''Shewanella'' is a facultative anaerobe, which means that it will use oxygen if available, but in the absence of oxygen can get rid of its electrons by dumping them on a wide range of alternate acceptors, of which nitrite is only one example. To handle the electron flow ''Shewanella'' uses a large number of promiscuous <scene name='Journal:JBIC:16/Cv/8'>c-heme</scene> containing electron transfer proteins. Indeed, ''Shewanella'' is exceptionally adept at producing c-heme proteins under fast-growth conditions, which many bacteria commonly used for large-scale laboratory gene expression, such as ''E. coli'', are incapable of unless they are first extensively reprogrammed genetically. Since ''Shewanella'' can be easily grown in the lab, and can naturally and easily produce c-hemes, it is an ideal host for generating large quantities of c-heme proteins such as ccNiR. | The ccNiR described here is produced by the ''Shewanella oneidensis'' bacterium, which is remarkable in its own right due to the large number of electron acceptors that it can utilize. ''Shewanella'' is a facultative anaerobe, which means that it will use oxygen if available, but in the absence of oxygen can get rid of its electrons by dumping them on a wide range of alternate acceptors, of which nitrite is only one example. To handle the electron flow ''Shewanella'' uses a large number of promiscuous <scene name='Journal:JBIC:16/Cv/8'>c-heme</scene> containing electron transfer proteins. Indeed, ''Shewanella'' is exceptionally adept at producing c-heme proteins under fast-growth conditions, which many bacteria commonly used for large-scale laboratory gene expression, such as ''E. coli'', are incapable of unless they are first extensively reprogrammed genetically. Since ''Shewanella'' can be easily grown in the lab, and can naturally and easily produce c-hemes, it is an ideal host for generating large quantities of c-heme proteins such as ccNiR. | ||
Revision as of 11:59, 13 August 2013
| |||||||||||
3D structures of nitric reductase
Cu-containing nitrite reductase with copper only
1nia, 1nib, 1nic, 1nid, 1nie, 1nif, 2nrd, 1kcb, 1rzp, 1rzq, 2bw4, 2bw5, 2avf – AcNIR + Cu – Achromobacter cycloclastes
2afn, 1aq8, 1as7, 2fjs, 2pp7, 2pp8, 3h4h, 3h56 - AfNIR + Cu – Alcaligenes faecalis
1ntd, 1npj, 1npn, 1zdq, 3h4f - AfNIR (mutant) + Cu
1ndr, 1ndt, 1bq5, 1hau, 1haw, 1oe1, 1oe3 - AxNIR + Cu – Achromobacter xylosoxidans
1oe2, 2jfc - AxNIR (mutant) + Cu
1mzz, 2dy2 - RsNIR + Cu – Rhodobacter sphaeroides
2dv6 - NIR + Cu – Hyphomicrobium denitrificans
Cu-containing nitrite reductase with variety of metals
1et5, 1et8 - AfNIR (mutant) + Zn + Cu
1et7 - AfNIR (mutant) + Cd + Cu
2vm3, 2vm4, 2vw4, 2vw6, 2vw7, 2vn3 - AxNIR + Zn + Cu
2bo0 - AxNIR (mutant) + Zn
2vmj - AxNIR + Zn
1gs6 - AxNIR (mutant) + Mg + Cu
1gs7, 1wae, 1wa0, 1wa1, 2bp0, 2bp8, 2xx0, 2xxf, 2xxg - AxNIR (mutant) + Zn + Cu
2zon - AxNIR + heme + Cu
1mzy, 1zv2, 2a3t - RsNIR + Mg + Cu
1n70 - RsNIR (mutant) + Mg + Cu
Cu-containing nitrite reductase binary complex
1as6, 1as8, 1sjm, 2ppc - AfNIR + NO2 + Cu
2e86 - AfNIR + N3 + Cu
1j9q, 1j9r, 1j9s, 1j9t, 1l9o, 1l9p, 1l9q, 1l9r, 1l9s, 1l9t - AfNIR (mutant) + NO2 + Cu
1zds, 2b08 - AfNIR (mutant) + acetamide + Cu
1snr - AfNIR + NO + Cu
2pp9 - AfNIR + NO3 + Cu
2ppa - AfNIR + N2O + Cu
2ppd, 2ppe, 2ppf - AfNIR (mutant) + NO + Cu
2p80 - AfNIR + pseudoazurin + Cu
1nds - AxNIR + NO2 + Cu
2xx1 - AxNIR (mutant) + NO2 + Cu
2xwz - AxNIR + NO + NO2 + Cu
1wa2 - AxNIR (mutant) + NO2 + Zn + Cu
2bwd, 2bwi - AcNIR + NO2 + Cu
2y1a - AcNIR + NO + Cu
2dws, 2dwt - RsNIR + NO2 + Cu
Heme-containing nitrite reductase
1aof, 1qks, 1hj4, 1hj5, 1h9x, 1hcm - PpNIR – Paracoccus pantotrophus
1gq1 - PpNIR (mutant)
1nir, 1bl9, 1n15, 1n50, 1n90 – PaNIR – Pseudomonas aeruginosa
1hzu - PaNIR (mutant)
1qdb - NIR – Sulfurospirillum deleyianum
1fs7, 1fs8 - WsNIR – Wolinella succinogenes
3bng - WsNIR (mutant)
1gu6, 2rdz, 3tor – EcNIR – Escherichia coli
2rf7 - EcNIR (mutant)
2jo6 – EcNIR small subunit – NMR
2jza - NIR small subunit – Pectobacterium atrosepticum - NMR
1oah – NIR – Desulfovibrio desulfuricans
2j7a - DvNIR – Desulfovibrio vulgaris
2ot4, 3gm6, 3fo3, 3sce, 3uu9 - TnNIR – Thioalkalivibrio nitratireducens
Heme-containing nitrite reductase binary complex
1dy7 - PpNIR + CO
1hj3 - PpNIR + O2
2e81 - WsNIR + NH2OH
2vr0 - DvNIR + HQNO inhibitor
3ziy, 4ax3 – RpNIR + Cu – Ralstonia pickettii
2yqb, 3zbm – RpNIR (mutant) + Cu
Heme-containing nitrite reductase binary complex with cyanide
1h9y - PpNIR + CN
1gjq - PaNIR + CN
1e2r - NIR + CN – Paracoccus denitrificans
Heme-containing nitrite reductase binary complex with nitric oxide
1nno - PaNIR + NO
1hzv - PaNIR (mutant) + NO
1aom, 1aoq - PpNIR + NO + NO2
Heme-containing nitrite reductase binary complex with nitrite
2e80 - WsNIR + NO2
3bnh - WsNIR (mutant) + NO
3d1i, 3rkh, 3owm - TnNIR + NO2
Heme-containing nitrite reductase binary complex with azide
2zo5 - TnNIR + N3
3s7w - TnNIR + NO2 + N3
1fs9 - WsNIR + N3
Heme-containing nitrite reductase binary complex with sulfite
3mmo - WsNIR + SO3
3bnj - WsNIR (mutant) + SO3
3lg1, 3lgq, 3f29, 3ttb - TnNIR + SO3
3l1t - EcNIR + SO3
Siroheme-containing nitrite reductase
2akj - NIR – spinach
3b0g, 3b0h - ToNIR – tobacco
3b0j, 3b0l, 3b0m, 3b0n - ToNIR (mutant)
- ↑ 1.0 1.1 1.2 Youngblut M, Judd ET, Srajer V, Sayyed B, Goelzer T, Elliott SJ, Schmidt M, Pacheco AA. Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system. J Biol Inorg Chem. 2012 Mar 2. PMID:22382353 doi:10.1007/s00775-012-0885-0