We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Glycogenin
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='3t7m' size='450' side='right' scene='' caption=''> | ||
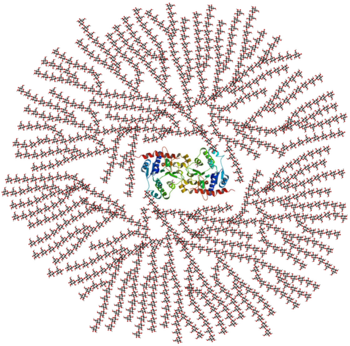
[[Image:Glycogen_structure.png|thumb|left|350x350px|alt=Alt text|Figure 1. A cross-sectional view of glycogen with the glycogenin dimer remaining covalently attached to the non-reducing end in the centre of the globule.]] | [[Image:Glycogen_structure.png|thumb|left|350x350px|alt=Alt text|Figure 1. A cross-sectional view of glycogen with the glycogenin dimer remaining covalently attached to the non-reducing end in the centre of the globule.]] | ||
| - | {{STRUCTURE_3t7m| PDB=3t7m | SIZE=400| SCENE= |right| CAPTION=Human glycogenin dimer complex with UDP, ethylene glycol and Mn+2 ion, [[3t7m]] }} | ||
'''Glycogenin''' (Glycogenin glucosyltransferase, [[EC]] 2.4.1.186) is a [[transferase]] responsible for the biosynthesis of glycogen; an important storage form of glucose in the body. It is a unique enzyme in that it is primer, substrate, catalyst, and product of its enzymatic reaction and extension process of glycogen biosynthesis. This is initiated by its ability to transfer glucose from UDP-glucose to form an oligosaccharide of glucose units that is covalently attached to itself at Tyr-194 in a multistep reaction mechanism <ref name="one"> PMID:12051921 </ref>. It is placed in glycosyltransferase family 8 becuase it contains highly conserved motifs that are common to glycosyltransferases such as lipopolysaccharide glucose and galactose transferases and galactinol synthases <ref name="two"> PMID:9345621 </ref>. | '''Glycogenin''' (Glycogenin glucosyltransferase, [[EC]] 2.4.1.186) is a [[transferase]] responsible for the biosynthesis of glycogen; an important storage form of glucose in the body. It is a unique enzyme in that it is primer, substrate, catalyst, and product of its enzymatic reaction and extension process of glycogen biosynthesis. This is initiated by its ability to transfer glucose from UDP-glucose to form an oligosaccharide of glucose units that is covalently attached to itself at Tyr-194 in a multistep reaction mechanism <ref name="one"> PMID:12051921 </ref>. It is placed in glycosyltransferase family 8 becuase it contains highly conserved motifs that are common to glycosyltransferases such as lipopolysaccharide glucose and galactose transferases and galactinol synthases <ref name="two"> PMID:9345621 </ref>. | ||
| Line 9: | Line 9: | ||
==Protein Function== | ==Protein Function== | ||
| - | + | <scene name='44/445344/Cv/1'>Figure 2. Glycogenin Monomer with UDP-glucose and Manganese</scene> (PDB entry [[1ll2]]). | |
| - | < | + | |
'''UDP-alpha-D-glucose + glycogenin <-> UDP + alpha-D-glucosylglycogenin''' | '''UDP-alpha-D-glucose + glycogenin <-> UDP + alpha-D-glucosylglycogenin''' | ||
| - | |||
The glycogenin binds <scene name='Sandbox_Reserved_343/Udp-glucose_and_manganese/2'>UDP-glucose and manganese</scene> at the Tyr-194 in <scene name='Sandbox_Reserved_343/Udp-glucose_and_manganese/3'>between the alpha/beta sandwich</scene> and then the enzyme is primed for extension by subsequent UDP-glucose additions for glycogen formation. The glucosyltransferase activity of glycogenin catalyzes the addition of subsequent UDP-glucose monomers to form a glucose polymer roughly 7 residues long. The reaction is then joined by the enzyme glycogen synthase which continues the α-1,4-glycosidic elongation of the glucose polymers, and glycogen branching enzyme that catalyzes α-1,6-glycosidic branch formation of the glycogen. The Mn2+ cation functions as a lewis acid to stabilize the UDP leaving group and help fascilitate the transfer from the Tyr-194 to another nucleophilic intermediate acceptor, Asp-162, in a dual-step nucleophilic SN1 substitution reaction <ref name="one" />. | The glycogenin binds <scene name='Sandbox_Reserved_343/Udp-glucose_and_manganese/2'>UDP-glucose and manganese</scene> at the Tyr-194 in <scene name='Sandbox_Reserved_343/Udp-glucose_and_manganese/3'>between the alpha/beta sandwich</scene> and then the enzyme is primed for extension by subsequent UDP-glucose additions for glycogen formation. The glucosyltransferase activity of glycogenin catalyzes the addition of subsequent UDP-glucose monomers to form a glucose polymer roughly 7 residues long. The reaction is then joined by the enzyme glycogen synthase which continues the α-1,4-glycosidic elongation of the glucose polymers, and glycogen branching enzyme that catalyzes α-1,6-glycosidic branch formation of the glycogen. The Mn2+ cation functions as a lewis acid to stabilize the UDP leaving group and help fascilitate the transfer from the Tyr-194 to another nucleophilic intermediate acceptor, Asp-162, in a dual-step nucleophilic SN1 substitution reaction <ref name="one" />. | ||
| Line 28: | Line 26: | ||
Glycogenin has been identified in two human isoforms. Glycogenin-1 is a 37kDa muscle isoform encoded for by the gene GYG1, whereas glycogenin-2 is the 66kDa liver isoform that is encoded by the gene GYG2 and expressed primarily in cardiac muscle <ref name="five"> PMID:20357282 </ref>. Mutations of the GYG1 gene results in a loss of the autoglycosylation capabilities of glycogenin for initiating glycogen synthesis in muscle, which leads to problems such as cardiac arrhythmia and muscle weakness due to depleted or abnormal storage of glycogen in heart and skeletal muscle <ref name="five" />. | Glycogenin has been identified in two human isoforms. Glycogenin-1 is a 37kDa muscle isoform encoded for by the gene GYG1, whereas glycogenin-2 is the 66kDa liver isoform that is encoded by the gene GYG2 and expressed primarily in cardiac muscle <ref name="five"> PMID:20357282 </ref>. Mutations of the GYG1 gene results in a loss of the autoglycosylation capabilities of glycogenin for initiating glycogen synthesis in muscle, which leads to problems such as cardiac arrhythmia and muscle weakness due to depleted or abnormal storage of glycogen in heart and skeletal muscle <ref name="five" />. | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
==3D structures of glycogenin== | ==3D structures of glycogenin== | ||
Revision as of 09:56, 21 August 2013
| |||||||||||
3D structures of glycogenin
Updated on 21-August-2013
3q4s - hGYG1 – human
1ll0, 1ll3, 3v8y – rGYG1 – rabbit
1zcu, 1zcv, 1zcy, 3usq, 3usr, 3v90 - rGYG1 (mutant)
Glycogenin complex with Mn+2 ion and UDP
3u2t - hGYG1 + Mn
3rmv – hGYG1 (mutant) + Mn + UDP
3rmw - hGYG1 (mutant) + Mn + UDP-glucose
3t7o - hGYG1 + Mn + UDP-glucose + glucose
3u2x - hGYG1 + Mn + UDP + glucose
3qvb, 3t7m, 3t7n - hGYG1 + Mn + UDP
3u2u - hGYG1 + Mn + UDP + maltotetraose
3u2v - hGYG1 + Mn + UDP + maltohexaose
3u2w - hGYG1 (mutant) + Mn + glucose
1zct, 3v8z – rGYG1 + Mn + UDP
1zdf, 1zdg, 3v91 - rGYG1 (mutant) + Mn + UDP-glucose
1ll2 - rGYG1 + Mn + UDP-glucose
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Gibbons BJ, Roach PJ, Hurley TD. Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J Mol Biol. 2002 May 31;319(2):463-77. PMID:12051921 doi:http://dx.doi.org/10.1016/S0022-2836(02)00305-4
- ↑ Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997 Oct;7(5):637-44. PMID:9345621
- ↑ Alonso MD, Lomako J, Lomako WM, Whelan WJ. A new look at the biogenesis of glycogen. FASEB J. 1995 Sep;9(12):1126-37. PMID:7672505
- ↑ 4.0 4.1 Shearer J, Wilson RJ, Battram DS, Richter EA, Robinson DL, Bakovic M, Graham TE. Increases in glycogenin and glycogenin mRNA accompany glycogen resynthesis in human skeletal muscle. Am J Physiol Endocrinol Metab. 2005 Sep;289(3):E508-14. Epub 2005 May 3. PMID:15870102 doi:10.1152/ajpendo.00100.2005
- ↑ 5.0 5.1 Moslemi AR, Lindberg C, Nilsson J, Tajsharghi H, Andersson B, Oldfors A. Glycogenin-1 deficiency and inactivated priming of glycogen synthesis. N Engl J Med. 2010 Apr 1;362(13):1203-10. PMID:20357282 doi:10.1056/NEJMoa0900661
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Kim Settle