Factor XIa
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='1zmn' size='450' side='right' scene='' caption=''> | <StructureSection load='1zmn' size='450' side='right' scene='' caption=''> | ||
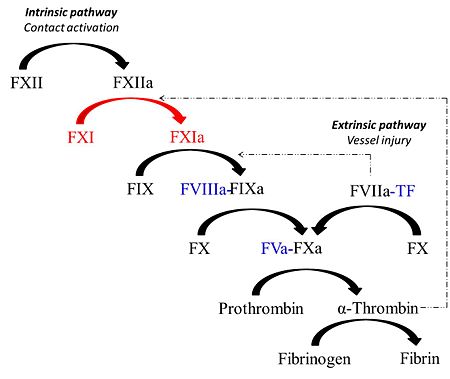
[[Image:Coag_cartoon.jpg |left| thumb |450px| Schematic representation of the coagulation response]] | [[Image:Coag_cartoon.jpg |left| thumb |450px| Schematic representation of the coagulation response]] | ||
| - | + | {{Clear}} | |
==Coagulation Factor XIa== | ==Coagulation Factor XIa== | ||
===Introduction=== | ===Introduction=== | ||
| Line 18: | Line 18: | ||
'''β-turn''' | '''β-turn''' | ||
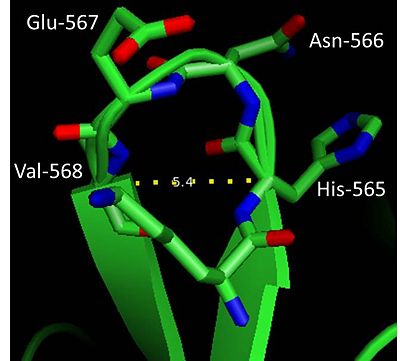
[[Image:Beta_turn.jpg | thumb |400px| β-turn in factor XIa]] | [[Image:Beta_turn.jpg | thumb |400px| β-turn in factor XIa]] | ||
| + | {{Clear}} | ||
The globular and compact nature of factor XIa as opposed to an elongated form (prevalent in vitamin K-dependent serine proteases) could in part be attributed to the abundance of β-turns in the protein. β-turns are characterized by a hydrogen bond involving carbonyl oxygen (C=O) of residue (i) and amide hydrogen (NH) of residue (i+3). The heavy chain has ~4 β-turns and one such β-turn (residues 566-568) is found in the light chain of factor XIa (see figure on the right). This β-turn based on distance between Cαi-Cαi+3 (5.4Å) and the measured dihedral angles: φ(i+1)=50.5°, ψ(i+1)=47.2° and φ(i+2)=90° and ψ(i+2)=15.8°could be classified as '''Type I′''' according to Hutchinson and Thornton (1994)<ref>PMID:7756980</ref>. | The globular and compact nature of factor XIa as opposed to an elongated form (prevalent in vitamin K-dependent serine proteases) could in part be attributed to the abundance of β-turns in the protein. β-turns are characterized by a hydrogen bond involving carbonyl oxygen (C=O) of residue (i) and amide hydrogen (NH) of residue (i+3). The heavy chain has ~4 β-turns and one such β-turn (residues 566-568) is found in the light chain of factor XIa (see figure on the right). This β-turn based on distance between Cαi-Cαi+3 (5.4Å) and the measured dihedral angles: φ(i+1)=50.5°, ψ(i+1)=47.2° and φ(i+2)=90° and ψ(i+2)=15.8°could be classified as '''Type I′''' according to Hutchinson and Thornton (1994)<ref>PMID:7756980</ref>. | ||
'''α-Helix''' | '''α-Helix''' | ||
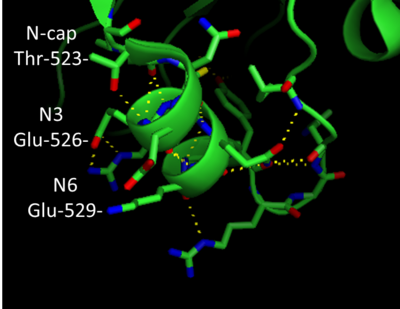
[[Image:Ncap.png | thumb |400px| N-terminal capping motif in factor XIa]] | [[Image:Ncap.png | thumb |400px| N-terminal capping motif in factor XIa]] | ||
| + | {{Clear}} | ||
In addition to β-sheets, factor XIa folds into a number of α-helices. The heavy chain region has ~8 helix repeats while about 5 helix repeats are present in the light chain region of the protein. Helices that are capped at either the N or C-terminals forming a capping motif. '''''Helix Capping Motif''''' display a unique hydrogen bonding pattern in addition to hydrophobic interactions. Factor XIa has an N-terminal capping motif in the light chain: residues 523-531 form an α-helix (see the figure on the right). Thr-532 is the Ncap and Glu-526 is found at the N3 position of the helix. The N-terminal capping motif shown in the figure (right hand side of page) appears to belong to the '''capping box''' classification. Thus the side chain of Thr-523 forms a H-bond with the backbone of the N3 however, H-bonding between the side chain of N3 and backbone of Ncap is absent. Next to the C-terminal Cys356 of the <scene name='Sandbox/A4_domain/2'>A4</scene> domain,factor XI assumes an interesting helical element the <scene name='Sandbox/310_helix_of_a4_domain/1'>3-10 helical structure</scene>. Originally classified as a type III turn, the <scene name='Sandbox/310_helix_of_a4_domain/1'>3-10 helical structure</scene> is tight and contains 3 residues (357-360) per turn. | In addition to β-sheets, factor XIa folds into a number of α-helices. The heavy chain region has ~8 helix repeats while about 5 helix repeats are present in the light chain region of the protein. Helices that are capped at either the N or C-terminals forming a capping motif. '''''Helix Capping Motif''''' display a unique hydrogen bonding pattern in addition to hydrophobic interactions. Factor XIa has an N-terminal capping motif in the light chain: residues 523-531 form an α-helix (see the figure on the right). Thr-532 is the Ncap and Glu-526 is found at the N3 position of the helix. The N-terminal capping motif shown in the figure (right hand side of page) appears to belong to the '''capping box''' classification. Thus the side chain of Thr-523 forms a H-bond with the backbone of the N3 however, H-bonding between the side chain of N3 and backbone of Ncap is absent. Next to the C-terminal Cys356 of the <scene name='Sandbox/A4_domain/2'>A4</scene> domain,factor XI assumes an interesting helical element the <scene name='Sandbox/310_helix_of_a4_domain/1'>3-10 helical structure</scene>. Originally classified as a type III turn, the <scene name='Sandbox/310_helix_of_a4_domain/1'>3-10 helical structure</scene> is tight and contains 3 residues (357-360) per turn. | ||
| Line 40: | Line 42: | ||
===Active Site Residues=== | ===Active Site Residues=== | ||
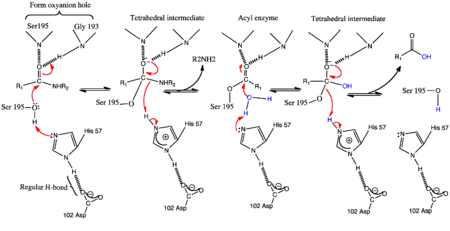
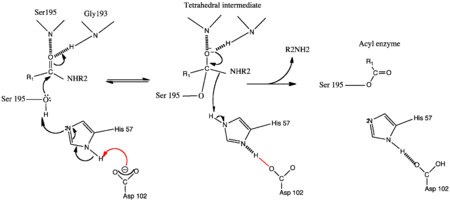
| - | [[Image:Genmechanism.png| thumb | | + | [[Image:Genmechanism.png|left| thumb |450px| The Conventional General base Mechanism of serine proteases]] |
| + | {{Clear}} | ||
Similar to other members of the serine protease family, factor XIa bears the two β-barrels topology unique to chymotrypsin-like proteases which are linked through a central loop. Thus cleavage at <scene name='Sandbox/Arg369-ile370/1'> Arg369-Ile370</scene> of the zymogen generates a functional enzyme. the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] residues Ser-557, Asp-462 and His-413 constitute the <scene name='Sandbox/Active_site/2'>active site</scene> of factor XIa. | Similar to other members of the serine protease family, factor XIa bears the two β-barrels topology unique to chymotrypsin-like proteases which are linked through a central loop. Thus cleavage at <scene name='Sandbox/Arg369-ile370/1'> Arg369-Ile370</scene> of the zymogen generates a functional enzyme. the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] residues Ser-557, Asp-462 and His-413 constitute the <scene name='Sandbox/Active_site/2'>active site</scene> of factor XIa. | ||
| Line 48: | Line 51: | ||
Thus, the Nε2 of His57 acts a the general base to enhance the nucleophilicity of the catalytic Ser195-OH by abstracting its proton. The acylation phase occurs with the attack of the carbonyl of the scissile bond generation the transition state intermediate. Also known as the tetrahedral intermediate, the negative oxygen ion in this structure is thought to be stabilized by the oxyanion hole formed by the amide backbone hydrogens of Ser195 and Gly193. The collapse of this intermediate involves the abstraction of the Nε2-H of His57 (general acid) by the leaving group P1' nitrogen of the substrate forming the first product and an acylenzyme intermediate<ref>PMID:332063</ref>. | Thus, the Nε2 of His57 acts a the general base to enhance the nucleophilicity of the catalytic Ser195-OH by abstracting its proton. The acylation phase occurs with the attack of the carbonyl of the scissile bond generation the transition state intermediate. Also known as the tetrahedral intermediate, the negative oxygen ion in this structure is thought to be stabilized by the oxyanion hole formed by the amide backbone hydrogens of Ser195 and Gly193. The collapse of this intermediate involves the abstraction of the Nε2-H of His57 (general acid) by the leaving group P1' nitrogen of the substrate forming the first product and an acylenzyme intermediate<ref>PMID:332063</ref>. | ||
| - | During the deacylation phase of the mechanism,water molecule is activated to act as the nucleophile by the abstration of a proton by the Nε2 of His57. The resulting nucleophilic OH anion attacks the carbonyl of the acylenzyme to generate another tetrahedral intermediate whose subsequent collapse releases the carboxylic half of the second product<ref>PMID:4372018</ref>.[[Image:ChargeRelay.png| thumb| | + | During the deacylation phase of the mechanism,water molecule is activated to act as the nucleophile by the abstration of a proton by the Nε2 of His57. The resulting nucleophilic OH anion attacks the carbonyl of the acylenzyme to generate another tetrahedral intermediate whose subsequent collapse releases the carboxylic half of the second product<ref>PMID:4372018</ref>. |
| - | + | [[Image:ChargeRelay.png|left| thumb|450px| The Charge Relay Mechanism of serine proteases]] | |
| + | {{Clear}} | ||
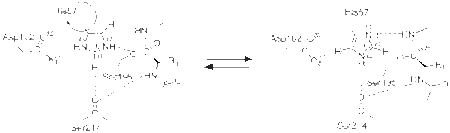
===The Charge Relay Mechanism of serine proteases=== | ===The Charge Relay Mechanism of serine proteases=== | ||
Blow and colleagues crystallized chymotrypsin in the presence of K2PtCL4 at a resolution of 5.8 Å and proposed a hydrogen bonding network between Asp102, His57 and Ser195<ref>PMID:5764436</ref>. Popularly known as the charge relay mechanism, Blow and colleagues proposed that, a two concerted proton transfers must occur in the transition state for the tetrahedral intermediate to be formed. Thus inaddition to His57, Asp102 acts a base to abstract a proton from the Nδ1-H of His57. On the contrary, later studies refuted the role of Asp102 as a base. Neutron diffraction studies<ref>PMID:7030393</ref> and NMR data<ref>PMID:31898</ref>revealed His57 as the only base and the presence of positive charge on the imidazole ring respectively. | Blow and colleagues crystallized chymotrypsin in the presence of K2PtCL4 at a resolution of 5.8 Å and proposed a hydrogen bonding network between Asp102, His57 and Ser195<ref>PMID:5764436</ref>. Popularly known as the charge relay mechanism, Blow and colleagues proposed that, a two concerted proton transfers must occur in the transition state for the tetrahedral intermediate to be formed. Thus inaddition to His57, Asp102 acts a base to abstract a proton from the Nδ1-H of His57. On the contrary, later studies refuted the role of Asp102 as a base. Neutron diffraction studies<ref>PMID:7030393</ref> and NMR data<ref>PMID:31898</ref>revealed His57 as the only base and the presence of positive charge on the imidazole ring respectively. | ||
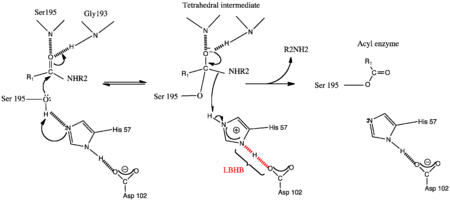
===The LBHB-mediated General Base Mechanism of serine proteases=== | ===The LBHB-mediated General Base Mechanism of serine proteases=== | ||
| - | NMR studies looking at the chemical shifts for the imadazonium ion (12ppm) in the tetrahedral intermediate and imidazole ring (11pm) in both the conventioanl general base and charge relay mechanisms respectively, were entirely different from the low field proton of His57 which was greater than 18ppm. This value is characteristic of a [http://en.wikipedia.org/wiki/Low-barrier_hydrogen_bond low barrier hydrogen bond (LBHB)].[[Image:LBHB.png| thumb | | + | NMR studies looking at the chemical shifts for the imadazonium ion (12ppm) in the tetrahedral intermediate and imidazole ring (11pm) in both the conventioanl general base and charge relay mechanisms respectively, were entirely different from the low field proton of His57 which was greater than 18ppm. This value is characteristic of a [http://en.wikipedia.org/wiki/Low-barrier_hydrogen_bond low barrier hydrogen bond (LBHB)]. |
| + | [[Image:LBHB.png|left| thumb |450px| The LBHB-mediated General base Mechanism of serine proteases]] | ||
| + | {{Clear}} | ||
| + | LBHB is a strong hydrogen bond formed between two heteroatoms in which the distance of the hydrogen from either the donor or acceptor is equivalent (i.e. <2,55Å) and the ΔpKa between heteroatoms sharing the H-bond is ~0<ref>PMID:7661899</ref>. | ||
| Line 60: | Line 67: | ||
===The His Flip Mechanism of Serine Proteases=== | ===The His Flip Mechanism of Serine Proteases=== | ||
| - | [[Image:His flip modified.gif| | + | [[Image:His flip modified.gif|left|thumb|450px| '''The His Flip mechanism of Serine proteases''']] |
| + | {{Clear}} | ||
Studies on the central role of His57 and its orientation as regards functioning as the general base and general acid, led to the proposal of the '''His Moving''' mechanism. Earlier kinetics studies using subtilisin as the model organism revealed that, whereas the kcat for ester hydrolysis in 50% N,N-dimethylformamide (DMF) remains unchanged, hydrolysis of amides was 2-fold lower. | Studies on the central role of His57 and its orientation as regards functioning as the general base and general acid, led to the proposal of the '''His Moving''' mechanism. Earlier kinetics studies using subtilisin as the model organism revealed that, whereas the kcat for ester hydrolysis in 50% N,N-dimethylformamide (DMF) remains unchanged, hydrolysis of amides was 2-fold lower. | ||
Revision as of 11:12, 26 August 2013
| |||||||||||
3D structures of Factor XI
Updated on 26-August-2013
Factor XI
2j8j, 2j8l – hFXIa A4 domain – human – NMR
2f83 - hFXI zymogen
Factor XI inhibitor complex
3bg8 – hFXIa + clavatadine
1zom, 1zsj, 1zsk, 1zlr, 1zmj, 1zml, 1zmn, 1zpz, 1zrk, 1ztj, 1ztk, 1ztl, 1zpb, 1zpc, 2fda, 1zsl, 1zhm, 1zhp, 1zhr, 3sor, 3sos - hFXI catalytic domain (mutant) + inhibitor
1zjd - hFXI catalytic domain (mutant) + Kunitz protease inhibitory domain
1xx9 - hFXI catalytic domain + ecotin (mutant)
1xxd, 1xxf - hFXI catalytic domain (mutant) + ecotin (mutant)
References
- ↑ Thompson RE, Mandle R Jr, Kaplan AP. Association of factor XI and high molecular weight kininogen in human plasma. J Clin Invest. 1977 Dec;60(6):1376-80. PMID:915004 doi:http://dx.doi.org/10.1172/JCI108898
- ↑ Baglia FA, Gailani D, Lopez JA, Walsh PN. Identification of a binding site for glycoprotein Ibalpha in the Apple 3 domain of factor XI. J Biol Chem. 2004 Oct 29;279(44):45470-6. Epub 2004 Aug 17. PMID:15317813 doi:10.1074/jbc.M406727200
- ↑ Herwald H, Jahnen-Dechent W, Alla SA, Hock J, Bouma BN, Muller-Esterl W. Mapping of the high molecular weight kininogen binding site of prekallikrein. Evidence for a discontinuous epitope formed by distinct segments of the prekallikrein heavy chain. J Biol Chem. 1993 Jul 5;268(19):14527-35. PMID:7686159
- ↑ Bouma BN, Griffin JH. Human blood coagulation factor XI. Purification, properties, and mechanism of activation by activated factor XII. J Biol Chem. 1977 Sep 25;252(18):6432-7. PMID:893417
- ↑ Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999 Nov 12;461(1-2):63-7. PMID:10561497
- ↑ Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999 Nov 12;461(1-2):63-7. PMID:10561497
- ↑ Hutchinson EG, Thornton JM. A revised set of potentials for beta-turn formation in proteins. Protein Sci. 1994 Dec;3(12):2207-16. PMID:7756980 doi:http://dx.doi.org/10.1002/pro.5560031206
- ↑ Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006 Jun;13(6):557-8. Epub 2006 May 14. PMID:16699514 doi:10.1038/nsmb1095
- ↑ Dorfman R, Walsh PN. Noncovalent interactions of the Apple 4 domain that mediate coagulation factor XI homodimerization. J Biol Chem. 2001 Mar 2;276(9):6429-38. Epub 2000 Nov 22. PMID:11092900 doi:10.1074/jbc.M010340200
- ↑ McMullen BA, Fujikawa K, Davie EW. Location of the disulfide bonds in human coagulation factor XI: the presence of tandem apple domains. Biochemistry. 1991 Feb 26;30(8):2056-60. PMID:1998667
- ↑ Imami K, Sugiyama N, Kyono Y, Tomita M, Ishihama Y. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column. Anal Sci. 2008 Jan;24(1):161-6. PMID:18187866
- ↑ Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009 Feb;8(2):651-61. PMID:19159218 doi:10.1021/pr8008012
- ↑ Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006 Jun;13(6):557-8. Epub 2006 May 14. PMID:16699514 doi:10.1038/nsmb1095
- ↑ Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003 Oct 2;425(6957):535-9. PMID:14523451 doi:10.1038/nature01962
- ↑ Sinha D, Marcinkiewicz M, Navaneetham D, Walsh PN. Macromolecular substrate-binding exosites on both the heavy and light chains of factor XIa mediate the formation of the Michaelis complex required for factor IX-activation. Biochemistry. 2007 Aug 28;46(34):9830-9. Epub 2007 Aug 4. PMID:17676929 doi:10.1021/bi062296c
- ↑ Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006 Jun;13(6):557-8. Epub 2006 May 14. PMID:16699514 doi:10.1038/nsmb1095
- ↑ Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331-58. PMID:332063 doi:http://dx.doi.org/10.1146/annurev.bi.46.070177.001555
- ↑ Bender ML, Killheffer JV. Chymotrypsins. CRC Crit Rev Biochem. 1973 Apr;1(2):149-99. PMID:4372018
- ↑ Blow DM, Birktoft JJ, Hartley BS. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature. 1969 Jan 25;221(5178):337-40. PMID:5764436
- ↑ Kossiakoff AA, Spencer SA. Direct determination of the protonation states of aspartic acid-102 and histidine-57 in the tetrahedral intermediate of the serine proteases: neutron structure of trypsin. Biochemistry. 1981 Oct 27;20(22):6462-74. PMID:7030393
- ↑ Markley JL, Ibanez IB. Zymogen activation in serine proteinases. Proton magnetic resonance pH titration studies of the two histidines of bovine chymotrypsinogen A and chymotrypsin Aalpha. Biochemistry. 1978 Oct 31;17(22):4627-40. PMID:31898
- ↑ Frey PA, Whitt SA, Tobin JB. A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science. 1994 Jun 24;264(5167):1927-30. PMID:7661899
- ↑ Frey PA, Whitt SA, Tobin JB. A low-barrier hydrogen bond in the catalytic triad of serine proteases. Science. 1994 Jun 24;264(5167):1927-30. PMID:7661899
- ↑ Henderson R. Catalytic activity of -chymotrypsin in which histidine-57 has been methylated. Biochem J. 1971 Aug;124(1):13-8. PMID:5126468
- ↑ Craik CS, Roczniak S, Largman C, Rutter WJ. The catalytic role of the active site aspartic acid in serine proteases. Science. 1987 Aug 21;237(4817):909-13. PMID:3303334
- ↑ Kidd RD, Sears P, Huang DH, Witte K, Wong CH, Farber GK. Breaking the low barrier hydrogen bond in a serine protease. Protein Sci. 1999 Feb;8(2):410-7. PMID:10048334
- ↑ Bachovchin WW. 15N NMR spectroscopy of hydrogen-bonding interactions in the active site of serine proteases: evidence for a moving histidine mechanism. Biochemistry. 1986 Nov 18;25(23):7751-9. PMID:3542033
- ↑ Kidd RD, Sears P, Huang DH, Witte K, Wong CH, Farber GK. Breaking the low barrier hydrogen bond in a serine protease. Protein Sci. 1999 Feb;8(2):410-7. PMID:10048334
- ↑ Bachovchin WW. 15N NMR spectroscopy of hydrogen-bonding interactions in the active site of serine proteases: evidence for a moving histidine mechanism. Biochemistry. 1986 Nov 18;25(23):7751-9. PMID:3542033
- ↑ Brady K, Wei AZ, Ringe D, Abeles RH. Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors. Biochemistry. 1990 Aug 21;29(33):7600-7. PMID:2271520
- ↑ Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, Almo SC. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001 Jan 30;40(4):853-60. PMID:11170405
- ↑ Ponczek MB, Gailani D, Doolittle RF. Evolution of the contact phase of vertebrate blood coagulation. J Thromb Haemost. 2008 Nov;6(11):1876-83. Epub 2008 Aug 28. PMID:18761718 doi:10.1111/j.1538-7836.2008.03143.x
- ↑ McMullen BA, Fujikawa K, Davie EW. Location of the disulfide bonds in human plasma prekallikrein: the presence of four novel apple domains in the amino-terminal portion of the molecule. Biochemistry. 1991 Feb 26;30(8):2050-6. PMID:1998666
- ↑ Ponczek MB, Gailani D, Doolittle RF. Evolution of the contact phase of vertebrate blood coagulation. J Thromb Haemost. 2008 Nov;6(11):1876-83. Epub 2008 Aug 28. PMID:18761718 doi:10.1111/j.1538-7836.2008.03143.x
- ↑ Saito H, Ratnoff OD, Bouma BN, Seligsohn U. Failure to detect variant (CRM+) plasma thromboplastin antecedent (factor XI) molecules in hereditary plasma thromboplastin antecedent deficiency: a study of 125 patients of several ethnic backgrounds. J Lab Clin Med. 1985 Dec;106(6):718-22. PMID:4067382
- ↑ 10706899
- ↑ Wang X, Cheng Q, Xu L, Feuerstein GZ, Hsu MY, Smith PL, Seiffert DA, Schumacher WA, Ogletree ML, Gailani D. Effects of factor IX or factor XI deficiency on ferric chloride-induced carotid artery occlusion in mice. J Thromb Haemost. 2005 Apr;3(4):695-702. Epub 2005 Feb 23. PMID:15733058 doi:10.1111/j.1538-7836.2005.01236.x
- ↑ Riley PW, Cheng H, Samuel D, Roder H, Walsh PN. Dimer dissociation and unfolding mechanism of coagulation factor XI apple 4 domain: spectroscopic and mutational analysis. J Mol Biol. 2007 Mar 23;367(2):558-73. Epub 2006 Dec 29. PMID:17257616 doi:10.1016/j.jmb.2006.12.066
- ↑ Kravtsov DV, Wu W, Meijers JC, Sun MF, Blinder MA, Dang TP, Wang H, Gailani D. Dominant factor XI deficiency caused by mutations in the factor XI catalytic domain. Blood. 2004 Jul 1;104(1):128-34. Epub 2004 Mar 16. PMID:15026311 doi:10.1182/blood-2003-10-3530
- ↑ Kravtsov DV, Wu W, Meijers JC, Sun MF, Blinder MA, Dang TP, Wang H, Gailani D. Dominant factor XI deficiency caused by mutations in the factor XI catalytic domain. Blood. 2004 Jul 1;104(1):128-34. Epub 2004 Mar 16. PMID:15026311 doi:10.1182/blood-2003-10-3530