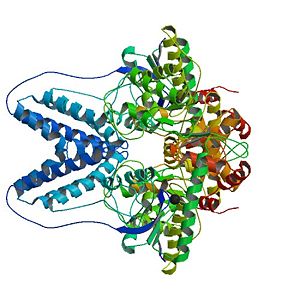
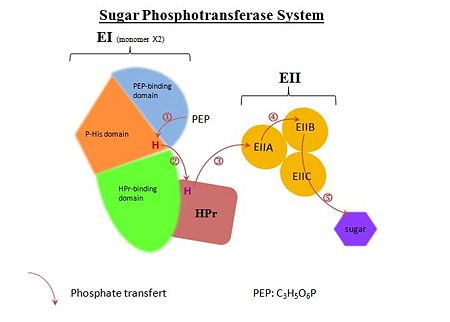
Enzyme I of the S. aureus PTS
From Proteopedia
| Line 35: | Line 35: | ||
-There is 920 Ǻ² of solvent accessible surface between the P-His with its phosphorylated His191 and the EIC domains with its PEP binding site. | -There is 920 Ǻ² of solvent accessible surface between the P-His with its phosphorylated His191 and the EIC domains with its PEP binding site. | ||
</StructureSection> | </StructureSection> | ||
| - | + | __NOTOC__ | |
== III.Staphylococcus aureus vs Escherichia coli == | == III.Staphylococcus aureus vs Escherichia coli == | ||
| - | |||
| - | |||
The X-ray structure of S. aureus EI (SaEI) has been recently solved but structures of other bacterias have been reported before, for instance E. coli EI structure (EcEI). Although SaEI and EcEI X-ray structures are nearly the same, some differences have been observed. | The X-ray structure of S. aureus EI (SaEI) has been recently solved but structures of other bacterias have been reported before, for instance E. coli EI structure (EcEI). Although SaEI and EcEI X-ray structures are nearly the same, some differences have been observed. | ||
| - | |||
[[Image:tableau SaEI EcEI.jpg]] | [[Image:tableau SaEI EcEI.jpg]] | ||
| - | |||
These two X-ray structures could actually be two possible conformations of the Enzyme I, corresponding to two steps of the phosphotransfer from PEP to HPr. EcEI structure correspond to the conformation II and SaEI structure correspond to the conformation I. | These two X-ray structures could actually be two possible conformations of the Enzyme I, corresponding to two steps of the phosphotransfer from PEP to HPr. EcEI structure correspond to the conformation II and SaEI structure correspond to the conformation I. | ||
| Line 50: | Line 46: | ||
== Conclusion: == | == Conclusion: == | ||
| - | |||
Knowing the structure of S. aureus EI allows a better comprehension of the PTS mechanism. But progresses could be still done such as the possible phenomena which induce the motions of the different domains. | Knowing the structure of S. aureus EI allows a better comprehension of the PTS mechanism. But progresses could be still done such as the possible phenomena which induce the motions of the different domains. | ||
| - | |||
== Reference == | == Reference == | ||
| Line 59: | Line 53: | ||
Oberholzer AE, Schneider P, Siebold C, Baumann U, Erni B. (2009).Crystal structure of enzyme I of the phosphoenolpyruvate:Sugar phosphotransferase system in the dephosphorylated state. J Biol Chem | Oberholzer AE, Schneider P, Siebold C, Baumann U, Erni B. (2009).Crystal structure of enzyme I of the phosphoenolpyruvate:Sugar phosphotransferase system in the dephosphorylated state. J Biol Chem | ||
[http://www.ncbi.nlm.nih.gov/pubmed/19801641?dopt=Abstract PMID: 19801641] | [http://www.ncbi.nlm.nih.gov/pubmed/19801641?dopt=Abstract PMID: 19801641] | ||
| - | |||
== 3D Structures of EI == | == 3D Structures of EI == | ||
Revision as of 08:52, 27 August 2013
| |||||||||||
III.Staphylococcus aureus vs Escherichia coli
The X-ray structure of S. aureus EI (SaEI) has been recently solved but structures of other bacterias have been reported before, for instance E. coli EI structure (EcEI). Although SaEI and EcEI X-ray structures are nearly the same, some differences have been observed.
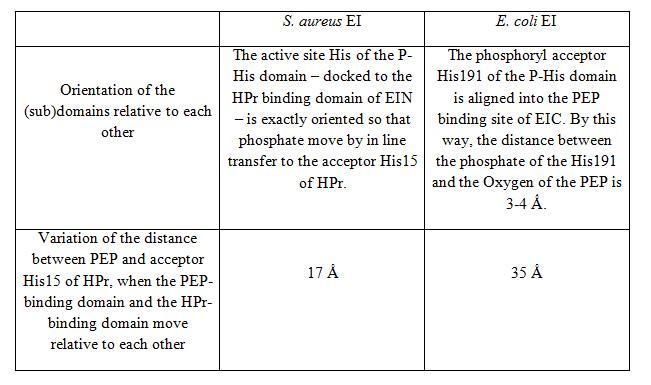 These two X-ray structures could actually be two possible conformations of the Enzyme I, corresponding to two steps of the phosphotransfer from PEP to HPr. EcEI structure correspond to the conformation II and SaEI structure correspond to the conformation I.
These two X-ray structures could actually be two possible conformations of the Enzyme I, corresponding to two steps of the phosphotransfer from PEP to HPr. EcEI structure correspond to the conformation II and SaEI structure correspond to the conformation I.
Thus, two modest rigid body motions can be described :
Conclusion:
Knowing the structure of S. aureus EI allows a better comprehension of the PTS mechanism. But progresses could be still done such as the possible phenomena which induce the motions of the different domains.
Reference
Oberholzer AE, Schneider P, Siebold C, Baumann U, Erni B. (2009).Crystal structure of enzyme I of the phosphoenolpyruvate:Sugar phosphotransferase system in the dephosphorylated state. J Biol Chem PMID: 19801641
3D Structures of EI
2wqd S. aureus EI phosphorylated structure
2hwg E. coli phosphorylated and Mg2+-oxalate bound structure
Links
Proteopedia page : Enzyme I of the Phosphoenolpyruvate:Sugar Phosphotransferase System (E. coli and S. carnosus)