We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox 128
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load= size= | + | <StructureSection load= size=550 side='right' scene='37/372728/Transpeptidase_in_rainbow/1'> |
==='''Introduction'''=== | ==='''Introduction'''=== | ||
Peptidoglycan transpeptidase (TP) also known as penicillin-binding proteins (PBP), are essential for bacterial cell wall synthesis and catalyze the cross-linking of peptidoglycan polymers during bacterial wall synthesis.[http://en.wikipedia.org/wiki/Beta-lactam_antibiotic Beta-lactam antibiotic], which includes the penicillins,cephalosporins,carbapenems, and the monobactam aztreonam, bind and irreversibly inhibit the active site of TP. The overuse and misuse of b-lactam antibiotics has led to strains of ''Staphylococcus aureus (S.aureus)'' that are resistant to all currently available b-lactams and are often susceptible to so-called "last resort antibiotics", such as vancomycin. | Peptidoglycan transpeptidase (TP) also known as penicillin-binding proteins (PBP), are essential for bacterial cell wall synthesis and catalyze the cross-linking of peptidoglycan polymers during bacterial wall synthesis.[http://en.wikipedia.org/wiki/Beta-lactam_antibiotic Beta-lactam antibiotic], which includes the penicillins,cephalosporins,carbapenems, and the monobactam aztreonam, bind and irreversibly inhibit the active site of TP. The overuse and misuse of b-lactam antibiotics has led to strains of ''Staphylococcus aureus (S.aureus)'' that are resistant to all currently available b-lactams and are often susceptible to so-called "last resort antibiotics", such as vancomycin. | ||
| + | |||
==='''Bacterial Cell Wall Structure'''=== | ==='''Bacterial Cell Wall Structure'''=== | ||
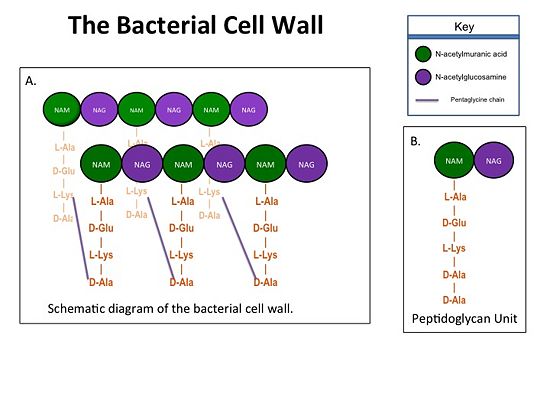
The bacterial cell wall is crucial for maintaining the structural integrity of bacteria and protects bacteria from osmotic stress and toxic compound. The cell wall is composed of peptidoglycan (Figure 2), and in Gram positive bacterial species (e.g. S. aureus) is many layers thick, while in Gram negative bacterial species (e.g. Escherichia coli) is only a few layers thick. The difference in the number of peptidoglycan layers accounts for the differential staining of these two groups of organisms. Peptidoglycan consists of a carbohydrate portion: alternating residues of [http://en.wikipedia.org/wiki/N-Acetylmuramic_acid N-acetylmuramic Acid (NAM)] and [http://en.wikipedia.org/wiki/N-Acetylglucosamine N-acetylglucosamine (NAG)] that polymerize to form long chains, and a protein portion: a pentapeptide chain that terminates with to D-alanines (D-Ala) residues. The pentapeptide chains are covalently bound to each NAM residue. Rows of peptidoglycan are cross-linked together with pentaglycine chains to form a "mesh-like" structure. This cross-linking reaction is catalyzed by TPs. | The bacterial cell wall is crucial for maintaining the structural integrity of bacteria and protects bacteria from osmotic stress and toxic compound. The cell wall is composed of peptidoglycan (Figure 2), and in Gram positive bacterial species (e.g. S. aureus) is many layers thick, while in Gram negative bacterial species (e.g. Escherichia coli) is only a few layers thick. The difference in the number of peptidoglycan layers accounts for the differential staining of these two groups of organisms. Peptidoglycan consists of a carbohydrate portion: alternating residues of [http://en.wikipedia.org/wiki/N-Acetylmuramic_acid N-acetylmuramic Acid (NAM)] and [http://en.wikipedia.org/wiki/N-Acetylglucosamine N-acetylglucosamine (NAG)] that polymerize to form long chains, and a protein portion: a pentapeptide chain that terminates with to D-alanines (D-Ala) residues. The pentapeptide chains are covalently bound to each NAM residue. Rows of peptidoglycan are cross-linked together with pentaglycine chains to form a "mesh-like" structure. This cross-linking reaction is catalyzed by TPs. | ||
[[Image:Cell Wall 7 30 2013.jpg|thumb|alt= Alt text| Figure 1.(A)Rows of Peptidoglycans forming a Bacterial Cell Wall (B)Peptidoglycan with D-Ala-D-Ala substrate |550px]] | [[Image:Cell Wall 7 30 2013.jpg|thumb|alt= Alt text| Figure 1.(A)Rows of Peptidoglycans forming a Bacterial Cell Wall (B)Peptidoglycan with D-Ala-D-Ala substrate |550px]] | ||
| + | |||
| + | |||
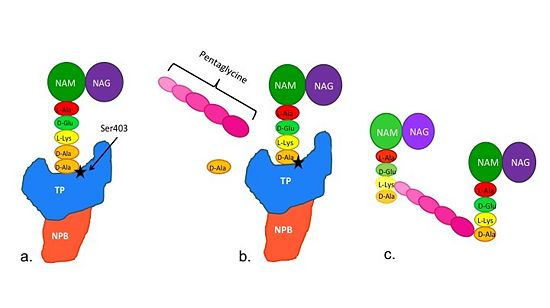
==='''Catalytic Mechanism of Action of Transpeptidases'''=== | ==='''Catalytic Mechanism of Action of Transpeptidases'''=== | ||
| - | [[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text|Figure 2. Schematic showing Catalytic Mechanism of PBP2a |550px]] | ||
(a)The D-Ala side-chain substrate accesses the TP active site. | (a)The D-Ala side-chain substrate accesses the TP active site. | ||
(b)The active site serine residue nucleophilically attacks the peptide bond between the terminal D-Ala residues. Terminal D-Ala residue exits the active site, and the remaining D-Ala residue forms a covalent bond with the active site serine residue to form an acyl-TP complex. | (b)The active site serine residue nucleophilically attacks the peptide bond between the terminal D-Ala residues. Terminal D-Ala residue exits the active site, and the remaining D-Ala residue forms a covalent bond with the active site serine residue to form an acyl-TP complex. | ||
(c)Subsequently, a pentaglycine chain enters the TP active site through nucleophillic attack forms a covalent bond with the D-Ala formerly bound to the active site serine residue. As a result, the TP site serine residue is regenerated. The entire process takes approximately 4 seconds. | (c)Subsequently, a pentaglycine chain enters the TP active site through nucleophillic attack forms a covalent bond with the D-Ala formerly bound to the active site serine residue. As a result, the TP site serine residue is regenerated. The entire process takes approximately 4 seconds. | ||
| + | [[Image:Schematic TP 3steps.jpg|thumb|alt= Alt text|Figure 2. Schematic showing Catalytic Mechanism of PBP2a |550px]] | ||
Revision as of 20:06, 30 September 2013
| |||||||||||