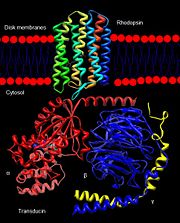
Rhodopsin
From Proteopedia
| Line 79: | Line 79: | ||
===Rhodopsin (Rn)=== | ===Rhodopsin (Rn)=== | ||
| - | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | |
[[3aym]], [[3ayn]], [[2z73]], [[2ziy]] – Rn + detergents + retinal – Flying squid<br /> | [[3aym]], [[3ayn]], [[2z73]], [[2ziy]] – Rn + detergents + retinal – Flying squid<br /> | ||
[[3am6]] – Rn-2 + cholesterol + retinal – ''Acetabularia acetabulum''<br /> | [[3am6]] – Rn-2 + cholesterol + retinal – ''Acetabularia acetabulum''<br /> | ||
| - | [[2x72]] – | + | [[2x72]] – bRn + detergents + retinal + guanine nucleotide-binding protein peptide – bovine<br /> |
| - | [[3qc9]] – | + | [[3qc9]] – bRn + Mg + ADP<br /> |
[[3oax]], [[3c9l]], [[2ped]], [[2i35]], [[2i36]], [[2i37]], [[1u19]], [[1gzm]], [[1l9h]], [[1hzx]] - bRn + detergents + retinal<br /> | [[3oax]], [[3c9l]], [[2ped]], [[2i35]], [[2i36]], [[2i37]], [[1u19]], [[1gzm]], [[1l9h]], [[1hzx]] - bRn + detergents + retinal<br /> | ||
[[3cap]] - bovine Rn + detergents<br /> | [[3cap]] - bovine Rn + detergents<br /> | ||
[[1jfp]], [[1f88]] - bRn + retinal<br /> | [[1jfp]], [[1f88]] - bRn + retinal<br /> | ||
| - | [[3c9m]] - | + | [[3c9m]] - bRn (mutant) + detergents + retinal<br /> |
| - | [[2j4y]] - | + | [[2j4y]], [[4bez]] - bRn (mutant) + detergents<br /> |
| - | [[3dqb]], [[4a4m]] - | + | [[3dqb]], [[4a4m]] - bRn + guanine nucleotide-binding protein peptide + detergents<br /> |
| - | [[1vqx]], [[1nzs]] – | + | [[4bey]] - bRn (mutant) + guanine nucleotide-binding protein peptide + detergents<br /> |
| - | [[1edx]] - | + | [[1vqx]], [[1nzs]] – bRn C terminal – NMR<BR /> |
| + | [[1edx]] - bRn N terminal – NMR<BR /> | ||
[[1eds]], [[1edv]], [[1edw]] – bRn intradiskal loop – NMR<BR /> | [[1eds]], [[1edv]], [[1edw]] – bRn intradiskal loop – NMR<BR /> | ||
| - | [[1fdf]] – | + | [[1fdf]] – bRn helix 7 - NMR<BR /> |
[[1xio]] – Rn - ''Anabaena''<br /> | [[1xio]] – Rn - ''Anabaena''<br /> | ||
[[1gu8]], [[1gue]], [[1h68]], [[1jgj]], [[3qap]], [[3qdc]] – NpRn II – ''Natronomonas pharaonis''<br /> | [[1gu8]], [[1gue]], [[1h68]], [[1jgj]], [[3qap]], [[3qdc]] – NpRn II – ''Natronomonas pharaonis''<br /> | ||
[[2ksy]] – NpRn II - NMR<br /> | [[2ksy]] – NpRn II - NMR<br /> | ||
| - | [[1h2s]], [[2f93]], [[2f95]] – NpRn II + Rn II transducer | + | [[1h2s]], [[2f93]], [[2f95]] – NpRn II + Rn II transducer<br /> |
| + | [[4gyc]] - NpRn II (mutant) + Rn II transducer<br /> | ||
===Metarhodopsin II=== | ===Metarhodopsin II=== | ||
| Line 118: | Line 120: | ||
[[1e12]], [[2jaf]] - HRn + detergents + retinal – ''Halobacterium salinarium''<br /> | [[1e12]], [[2jaf]] - HRn + detergents + retinal – ''Halobacterium salinarium''<br /> | ||
[[2jag]] - HRn (mutant) + detergents + retinal<br /> | [[2jag]] - HRn (mutant) + detergents + retinal<br /> | ||
| - | [[3a7k]], [[3abw]], [[3qbg]], [[3qbi]], [[3qbk]], [[3qbl]] – NpRn + detergents + retinal | + | [[3a7k]], [[3abw]], [[3qbg]], [[3qbi]], [[3qbk]], [[3qbl]], [[3vvk]] – NpRn + detergents + retinal |
===Archaerhodopsin=== | ===Archaerhodopsin=== | ||
| Line 128: | Line 130: | ||
===Proteorhodopsin=== | ===Proteorhodopsin=== | ||
| - | [[2l6x]] - | + | [[2l6x]] - GpPRn + detergents + retinal – ''Gamma proteobacterium'' |
| + | [[4kly]], [[4knf]] - GpPRn + retinal<br /> | ||
| + | [[4jq6]] – PRn + retinal + lipid<br /> | ||
===Xanthorhodopsin=== | ===Xanthorhodopsin=== | ||
[[3ddl]] - XRn + detergents + retinal – ''Salinibacter ruber'' | [[3ddl]] - XRn + detergents + retinal – ''Salinibacter ruber'' | ||
| + | |||
| + | ===Deltarhodopsin=== | ||
| + | |||
| + | [[4fbz]] - Rn + bacterioruberin + retinal – ''Haloterrigena thermotolerans''<br /> | ||
==References== | ==References== | ||
Revision as of 10:09, 13 October 2013
| |||||||||||
3D structures of rhodopsin
Rhodopsin (Rn)
Updated on 13-October-2013
3aym, 3ayn, 2z73, 2ziy – Rn + detergents + retinal – Flying squid
3am6 – Rn-2 + cholesterol + retinal – Acetabularia acetabulum
2x72 – bRn + detergents + retinal + guanine nucleotide-binding protein peptide – bovine
3qc9 – bRn + Mg + ADP
3oax, 3c9l, 2ped, 2i35, 2i36, 2i37, 1u19, 1gzm, 1l9h, 1hzx - bRn + detergents + retinal
3cap - bovine Rn + detergents
1jfp, 1f88 - bRn + retinal
3c9m - bRn (mutant) + detergents + retinal
2j4y, 4bez - bRn (mutant) + detergents
3dqb, 4a4m - bRn + guanine nucleotide-binding protein peptide + detergents
4bey - bRn (mutant) + guanine nucleotide-binding protein peptide + detergents
1vqx, 1nzs – bRn C terminal – NMR
1edx - bRn N terminal – NMR
1eds, 1edv, 1edw – bRn intradiskal loop – NMR
1fdf – bRn helix 7 - NMR
1xio – Rn - Anabaena
1gu8, 1gue, 1h68, 1jgj, 3qap, 3qdc – NpRn II – Natronomonas pharaonis
2ksy – NpRn II - NMR
1h2s, 2f93, 2f95 – NpRn II + Rn II transducer
4gyc - NpRn II (mutant) + Rn II transducer
Metarhodopsin II
3pqr – bMRn + guanine nucleotide-binding protein peptide + detergents + retinal
3pxo - bMRn + detergents + retinal
1ln6 - bMRn + retinal
Bathorhodopsin
2g87 - bBRn + detergents + retinal
Lumirhodopsin
2hpy - bLRn + detergents + retinal
Halorhodopsin
1e12, 2jaf - HRn + detergents + retinal – Halobacterium salinarium
2jag - HRn (mutant) + detergents + retinal
3a7k, 3abw, 3qbg, 3qbi, 3qbk, 3qbl, 3vvk – NpRn + detergents + retinal
Archaerhodopsin
1uaz – HRn-1 + retinal – Halobacterium
1vgo, 2z55 - HRn-2 + retinal
2ei4 - HRn-2 + retinal + bacterioruberin
Proteorhodopsin
2l6x - GpPRn + detergents + retinal – Gamma proteobacterium
4kly, 4knf - GpPRn + retinal
4jq6 – PRn + retinal + lipid
Xanthorhodopsin
3ddl - XRn + detergents + retinal – Salinibacter ruber
Deltarhodopsin
4fbz - Rn + bacterioruberin + retinal – Haloterrigena thermotolerans
References
- ↑ Hornak V, Ahuja S, Eilers M, Goncalves JA, Sheves M, Reeves PJ, Smith SO. Light activation of rhodopsin: insights from molecular dynamics simulations guided by solid-state NMR distance restraints. J Mol Biol. 2010 Feb 26;396(3):510-27. Epub 2009 Dec 11. PMID:20004206 doi:10.1016/j.jmb.2009.12.003
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr Opin Cell Biol. 2002 Apr;14(2):189-95. PMID:11891118
- ↑ 3.0 3.1 3.2 3.3 Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004 Jul;103(1):21-80. PMID:15251227 doi:10.1016/j.pharmthera.2004.05.002
- ↑ Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol Endocrinol. 2010 Jan;24(1):261-74. Epub 2009 Dec 17. PMID:20019124 doi:10.1210/me.2009-0473
- ↑ 5.0 5.1 5.2 Meng EC, Bourne HR. Receptor activation: what does the rhodopsin structure tell us? Trends Pharmacol Sci. 2001 Nov;22(11):587-93. PMID:11698103
- ↑ 6.0 6.1 Shieh T, Han M, Sakmar TP, Smith SO. The steric trigger in rhodopsin activation. J Mol Biol. 1997 Jun 13;269(3):373-84. PMID:9199406 doi:10.1006/jmbi.1997.1035
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 7.8 Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001 May;26(5):318-24. PMID:11343925
- ↑ 8.0 8.1 Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004 Sep 10;342(2):571-83. PMID:15327956 doi:10.1016/j.jmb.2004.07.044
- ↑ 9.0 9.1 Janz JM, Farrens DL. Assessing structural elements that influence Schiff base stability: mutants E113Q and D190N destabilize rhodopsin through different mechanisms. Vision Res. 2003 Dec;43(28):2991-3002. PMID:14611935
- ↑ 10.0 10.1 10.2 Kisselev OG. Focus on molecules: rhodopsin. Exp Eye Res. 2005 Oct;81(4):366-7. PMID:16051215 doi:10.1016/j.exer.2005.06.018
- ↑ 11.0 11.1 11.2 Verhoeven MA, Bovee-Geurts PH, de Groot HJ, Lugtenburg J, DeGrip WJ. Methyl substituents at the 11 or 12 position of retinal profoundly and differentially affect photochemistry and signalling activity of rhodopsin. J Mol Biol. 2006 Oct 13;363(1):98-113. Epub 2006 Jul 28. PMID:16962138 doi:10.1016/j.jmb.2006.07.039
- ↑ 12.0 12.1 12.2 12.3 Morris MB, Dastmalchi S, Church WB. Rhodopsin: structure, signal transduction and oligomerisation. Int J Biochem Cell Biol. 2009 Apr;41(4):721-4. Epub 2008 Aug 3. PMID:18692154 doi:10.1016/j.biocel.2008.04.025
- ↑ 13.0 13.1 13.2 13.3 13.4 Nelson, D., and Cox, M. Lehninger Principles of Biochemistry. 2008. 5th edition. W. H. Freeman and Company, New York, New York, USA. pp. 462-465.
- ↑ Hurley JB, Spencer M, Niemi GA. Rhodopsin phosphorylation and its role in photoreceptor function. Vision Res. 1998 May;38(10):1341-52. PMID:9667002
- ↑ 15.0 15.1 15.2 Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008 Jul 10;454(7201):183-7. Epub 2008 Jun 18. PMID:18563085 doi:10.1038/nature07063
- ↑ 16.0 16.1 Surya A, Knox BE. Enhancement of opsin activity by all-trans-retinal. Exp Eye Res. 1998 May;66(5):599-603. PMID:9628807 doi:10.1006/exer.1997.0453
See Also
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Wayne Decatur, Jaime Prilusky, Joel L. Sussman, Cinting Lim