This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2pfu
From Proteopedia
(New page: 200px<br /><applet load="2pfu" size="350" color="white" frame="true" align="right" spinBox="true" caption="2pfu" /> '''NMR strcuture determination of the periplasm...) |
|||
| Line 4: | Line 4: | ||
==Overview== | ==Overview== | ||
| - | The transport of iron complexes through outer membrane transporters from | + | The transport of iron complexes through outer membrane transporters from Gram-negative bacteria is highly dependent on the TonB system. Together, the three components of the system, TonB, ExbB and ExbD, energize the transport of iron complexes through the outer membrane by utilizing the proton motive force across the cytoplasmic membrane. The three-dimensional (3D) structure of the periplasmic domain of TonB has previously been determined. However, no detailed structural information for the other two components of the TonB system is currently available and their role in the iron-uptake process is not yet clearly understood. ExbD from Escherichia coli contains 141 residues distributed in three domains: a small N-terminal cytoplasmic region, a single transmembrane helix and a C-terminal periplasmic domain. Here we describe the first well-defined solution structure of the periplasmic domain of ExbD (residues 44-141) solved by multidimensional nuclear magnetic resonance (NMR) spectroscopy. The monomeric structure presents three clearly distinct regions: an N-terminal flexible tail (residues 44-63), a well-defined folded region (residues 64-133) followed by a small C-terminal flexible region (residues 134-141). The folded region is formed by two alpha-helices that are located on one side of a single beta-sheet. The central beta-sheet is composed of five beta-strands, with a mixed parallel and antiparallel arrangement. Unexpectedly, this fold closely resembles that found in the C-terminal lobe of the siderophore-binding proteins FhuD and CeuE. The ExbD periplasmic domain has a strong tendency to aggregate in vitro and 3D-TROSY (transverse relaxation optimized spectroscopy) NMR experiments of the deuterated protein indicate that the multimeric protein has nearly identical secondary structure to that of the monomeric form. Chemical shift perturbation studies suggest that the Glu-Pro region (residues 70-83) of TonB can bind weakly to the surface and the flexible C-terminal region of ExbD. At the same time the Lys-Pro region (residues 84-102) and the folded C-terminal domain (residues 150-239) of TonB do not show significant binding to ExbD, suggesting that the main interactions forming the TonB complex occur in the cytoplasmic membrane. |
==About this Structure== | ==About this Structure== | ||
| Line 10: | Line 10: | ||
==Reference== | ==Reference== | ||
| - | The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins., Garcia-Herrero A, Peacock RS, Howard SP, Vogel HJ, Mol Microbiol. 2007 Oct 10 | + | The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins., Garcia-Herrero A, Peacock RS, Howard SP, Vogel HJ, Mol Microbiol. 2007 Nov;66(4):872-89. Epub 2007 Oct 10. PMID:[http://ispc.weizmann.ac.il//pmbin/getpm?pmid=17927700 17927700] |
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
[[Category: Garcia-Herrero, A.]] | [[Category: Garcia-Herrero, A.]] | ||
| - | [[Category: Howard, P | + | [[Category: Howard, P S.]] |
| - | [[Category: Peacock, S | + | [[Category: Peacock, S R.]] |
| - | [[Category: Vogel, H | + | [[Category: Vogel, H J.]] |
[[Category: exbd]] | [[Category: exbd]] | ||
[[Category: periplasmic domain]] | [[Category: periplasmic domain]] | ||
| Line 23: | Line 23: | ||
[[Category: transport protein]] | [[Category: transport protein]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 18:28:59 2008'' |
Revision as of 16:29, 21 February 2008
|
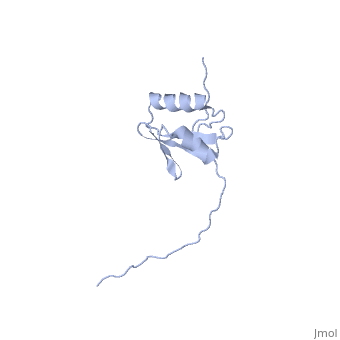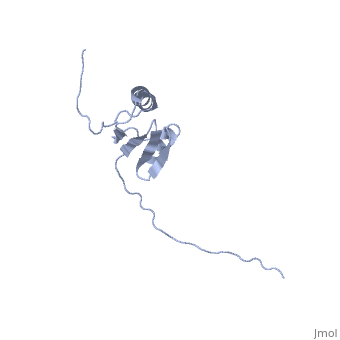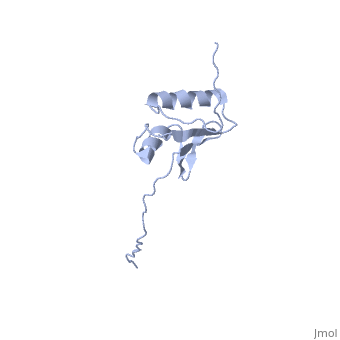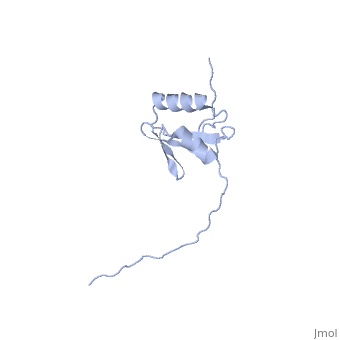
NMR strcuture determination of the periplasmic domain of ExbD from E.coli
Overview
The transport of iron complexes through outer membrane transporters from Gram-negative bacteria is highly dependent on the TonB system. Together, the three components of the system, TonB, ExbB and ExbD, energize the transport of iron complexes through the outer membrane by utilizing the proton motive force across the cytoplasmic membrane. The three-dimensional (3D) structure of the periplasmic domain of TonB has previously been determined. However, no detailed structural information for the other two components of the TonB system is currently available and their role in the iron-uptake process is not yet clearly understood. ExbD from Escherichia coli contains 141 residues distributed in three domains: a small N-terminal cytoplasmic region, a single transmembrane helix and a C-terminal periplasmic domain. Here we describe the first well-defined solution structure of the periplasmic domain of ExbD (residues 44-141) solved by multidimensional nuclear magnetic resonance (NMR) spectroscopy. The monomeric structure presents three clearly distinct regions: an N-terminal flexible tail (residues 44-63), a well-defined folded region (residues 64-133) followed by a small C-terminal flexible region (residues 134-141). The folded region is formed by two alpha-helices that are located on one side of a single beta-sheet. The central beta-sheet is composed of five beta-strands, with a mixed parallel and antiparallel arrangement. Unexpectedly, this fold closely resembles that found in the C-terminal lobe of the siderophore-binding proteins FhuD and CeuE. The ExbD periplasmic domain has a strong tendency to aggregate in vitro and 3D-TROSY (transverse relaxation optimized spectroscopy) NMR experiments of the deuterated protein indicate that the multimeric protein has nearly identical secondary structure to that of the monomeric form. Chemical shift perturbation studies suggest that the Glu-Pro region (residues 70-83) of TonB can bind weakly to the surface and the flexible C-terminal region of ExbD. At the same time the Lys-Pro region (residues 84-102) and the folded C-terminal domain (residues 150-239) of TonB do not show significant binding to ExbD, suggesting that the main interactions forming the TonB complex occur in the cytoplasmic membrane.
About this Structure
2PFU is a Single protein structure of sequence from Escherichia coli. Full crystallographic information is available from OCA.
Reference
The solution structure of the periplasmic domain of the TonB system ExbD protein reveals an unexpected structural homology with siderophore-binding proteins., Garcia-Herrero A, Peacock RS, Howard SP, Vogel HJ, Mol Microbiol. 2007 Nov;66(4):872-89. Epub 2007 Oct 10. PMID:17927700
Page seeded by OCA on Thu Feb 21 18:28:59 2008