We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Single stranded binding protein
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
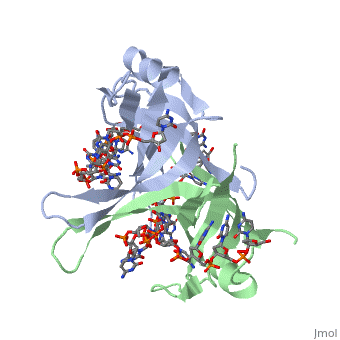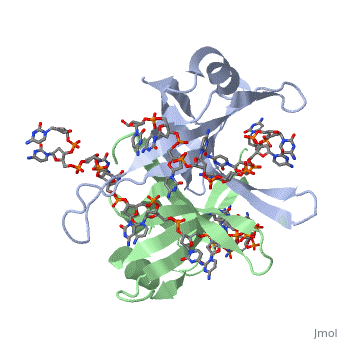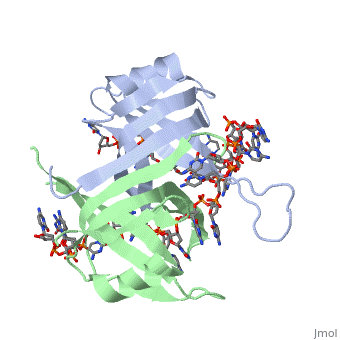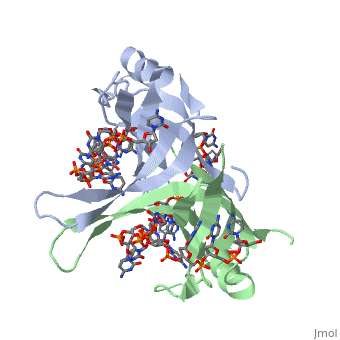
modification of arginine, cysteine, or tyrosine residues had no effect on binding of SSB to | modification of arginine, cysteine, or tyrosine residues had no effect on binding of SSB to | ||
DNA, whereas modification of either lysine residues (with acetic anhydride) or tryptophan | DNA, whereas modification of either lysine residues (with acetic anhydride) or tryptophan | ||
| - | residues (with N-bromosuccinimide) led to complete loss of binding activity | + | residues (with N-bromosuccinimide) led to complete loss of binding activity <ref>PMID: 2087220</ref>. |
The two tryptophan residues involved in DNA binding are Trp40 and Trp54, which was | The two tryptophan residues involved in DNA binding are Trp40 and Trp54, which was | ||
determined by mutagenesis. One more binding site was determined by site-specific mutagenesis. | determined by mutagenesis. One more binding site was determined by site-specific mutagenesis. | ||
Revision as of 15:08, 1 November 2013
Sandbox Single Stranded DNA-Binding Protein (SSB)
| |||||||||||
| |||||||||||
See Also
References
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
Proteopedia Page Contributors and Editors (what is this?)
Refayat Ahsen, Rachel Craig, Michal Harel, Alexander Berchansky