Single stranded binding protein
From Proteopedia
| Line 1: | Line 1: | ||
===Sandbox Single Stranded DNA-Binding Protein (SSB)=== | ===Sandbox Single Stranded DNA-Binding Protein (SSB)=== | ||
'''Single-stranded DNA-binding protein''', or SSB, binds to single-stranded regions of DNA in order to prevent premature annealing, to protect the single-stranded DNA from being digested by nucleases, and to remove secondary structure from the DNA to allow other enzymes to function effectively upon it. Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination and repair. As well as stabilizing this single-stranded DNA, SSB proteins bind to and modulate the function of numerous proteins involved in all of these processes. | '''Single-stranded DNA-binding protein''', or SSB, binds to single-stranded regions of DNA in order to prevent premature annealing, to protect the single-stranded DNA from being digested by nucleases, and to remove secondary structure from the DNA to allow other enzymes to function effectively upon it. Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination and repair. As well as stabilizing this single-stranded DNA, SSB proteins bind to and modulate the function of numerous proteins involved in all of these processes. | ||
| - | == | + | ==Overview== |
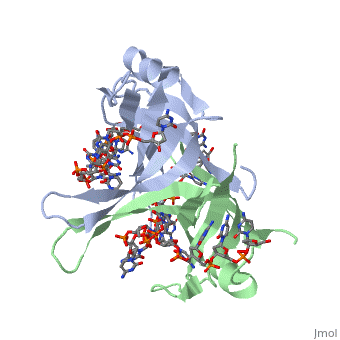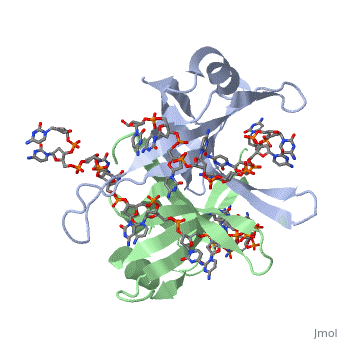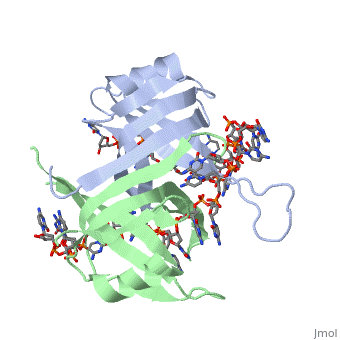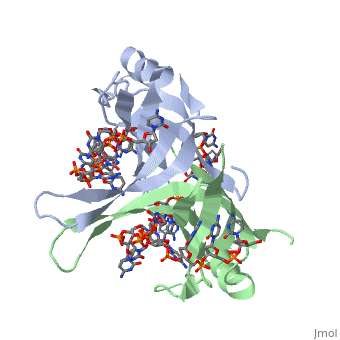
<StructureSection load='1eyg' size='400' side='right' frame='true' caption='Structure of Single Stranded DNA-Binding Protein bound to ssDNA (PDB entry [[1eyg]])' scene=''> | <StructureSection load='1eyg' size='400' side='right' frame='true' caption='Structure of Single Stranded DNA-Binding Protein bound to ssDNA (PDB entry [[1eyg]])' scene=''> | ||
| Line 11: | Line 11: | ||
==Structure== | ==Structure== | ||
| - | <StructureSection load='2vw9' size='400' side=' | + | <StructureSection load='2vw9' size='400' side='left' frame='true' caption='Structure of Single Stranded DNA-Binding Protein from Helicobacter Pylori bound to ssDNA (PDB entry [[2vw9]])' scene=''> |
SSB consists is a homotetramer that has a DNA binding domain which binds to a single strand of DNA. The tetramers consist of α-helices, β-sheets, and random coils. Each subunit contains an <scene name='56/566528/Alpha_helices/3'>α-helix</scene> and several <scene name='56/566528/Beta_sheets/1'>β-sheets</scene>. The secondary structure also includes a NH2 terminus, which consists of multiple positively charged amino acids. The DNA-binding domain lies within 115 amino acid residues from this terminus <ref>PMID: 2087220</ref>. The COOH terminus includes many acidic amino acids. | SSB consists is a homotetramer that has a DNA binding domain which binds to a single strand of DNA. The tetramers consist of α-helices, β-sheets, and random coils. Each subunit contains an <scene name='56/566528/Alpha_helices/3'>α-helix</scene> and several <scene name='56/566528/Beta_sheets/1'>β-sheets</scene>. The secondary structure also includes a NH2 terminus, which consists of multiple positively charged amino acids. The DNA-binding domain lies within 115 amino acid residues from this terminus <ref>PMID: 2087220</ref>. The COOH terminus includes many acidic amino acids. | ||
| Line 33: | Line 33: | ||
and a protein n, which is used to help synthesize RNA primers for the lagging strand. | and a protein n, which is used to help synthesize RNA primers for the lagging strand. | ||
</StructureSection> | </StructureSection> | ||
| + | |||
==Binding Interactions between DNA and SSB of ''E. coli''== | ==Binding Interactions between DNA and SSB of ''E. coli''== | ||
| - | <StructureSection load='1qvc' size='400' side=' | + | |
| + | <StructureSection load='1qvc' size='400' side='right' frame='true' caption='Structure of Single Stranded DNA-Binding Protein from E. coli (PDB entry [[1qvc]])' scene=''> | ||
Phe60 is an important DNA binding site. It has been shown to be the site for cross-linking. | Phe60 is an important DNA binding site. It has been shown to be the site for cross-linking. | ||
<scene name='56/566528/Tryptophan_residues/1'>Tryptophan</scene> and Lysine residues | <scene name='56/566528/Tryptophan_residues/1'>Tryptophan</scene> and Lysine residues | ||
Revision as of 02:25, 2 November 2013
Contents |
Sandbox Single Stranded DNA-Binding Protein (SSB)
Single-stranded DNA-binding protein, or SSB, binds to single-stranded regions of DNA in order to prevent premature annealing, to protect the single-stranded DNA from being digested by nucleases, and to remove secondary structure from the DNA to allow other enzymes to function effectively upon it. Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination and repair. As well as stabilizing this single-stranded DNA, SSB proteins bind to and modulate the function of numerous proteins involved in all of these processes.
Overview
| |||||||||||
Structure
| |||||||||||
Binding Interactions between DNA and SSB of E. coli
| |||||||||||
See Also
References
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
Proteopedia Page Contributors and Editors (what is this?)
Refayat Ahsen, Rachel Craig, Michal Harel, Alexander Berchansky