Glycogen synthase kinase 3
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='450' side='right' scene='Journal:JBIC:2/Opening/1' caption='Crystal Structure of Glycogen Synthase Kinase 3ß bound to Anticancer Ruthenium Complex'> | |
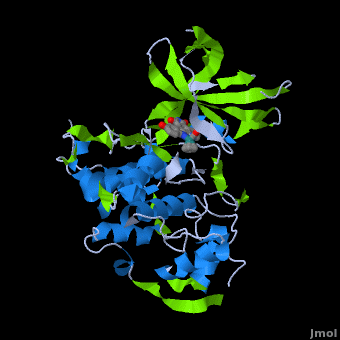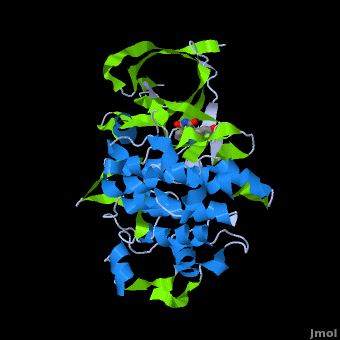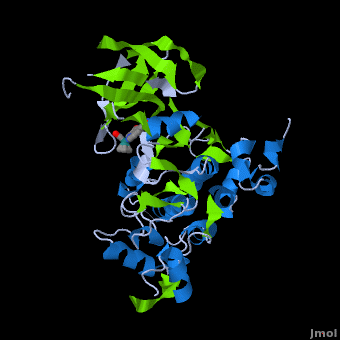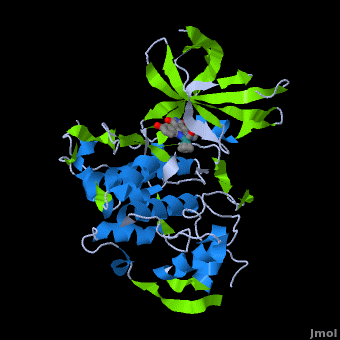
'''Glycogen synthase kinase 3''' (GSK-3) is a serine/threonine protein kinase. GSK-3 is active in a number of intracellular signaling pathways. GSK-3 regulates glycogen synthase as well as other proteins. GSK-3 inhibition is studied as a therapeutic target in diseases like Alzheimer, diabetes, bipolar disorder and some cancers. | '''Glycogen synthase kinase 3''' (GSK-3) is a serine/threonine protein kinase. GSK-3 is active in a number of intracellular signaling pathways. GSK-3 regulates glycogen synthase as well as other proteins. GSK-3 inhibition is studied as a therapeutic target in diseases like Alzheimer, diabetes, bipolar disorder and some cancers. | ||
| + | |||
| + | '''Structure of Anticancer Ruthenium Half-Sandwich Complex Bound to Glycogen Synthase Kinase 3ß <ref>DOI 10.1007/s00775-010-0699-x</ref>''' | ||
| + | A crystal structure of an <scene name='Journal:JBIC:2/Half_sandwich_complex_no_bonds/1'>organometallic half-sandwich ruthenium complex </scene>bound to the protein kinase glycogen synthase kinase 3ß (GSK-3ß) has been determined and reveals that the inhibitor binds to the <scene name='Journal:JBIC:2/Atp_binding_site2/2'>ATP binding site</scene> via an induced fit mechanism utlizing several <scene name='Journal:JBIC:2/Half_sandwich_complex/3'>hydrogen bonds</scene> and <scene name='Journal:JBIC:2/Half_sandwich_hydrophobic_stic/1'>hydrophobic interactions</scene>. Importantly, the metal is not involved in any direct interaction with the protein kinase but fulfills a purely structural role. The unique, bulky molecular structure of the half-sandwich complex with the CO-ligand oriented perpendicular to the pyridocarbazole heterocycle allows the complex to stretch the whole distance <scene name='Journal:JBIC:2/Half_sandwich_hydrophobic/5'>sandwiched between the faces of the N- and C-terminal lobes</scene> and to interact tightly with <scene name='Journal:JBIC:2/Glycine_rich_loop2/4'>the flexible glycine-rich loop</scene>. Although this complex is a conventional ATP-competitive binder, the unique shape of the complex allows novel interactions with the glycine-rich loop which are crucial for binding potency and selectivity. It can be hypothesized that coordination spheres which present other ligands towards the glycine-rich loop might display completely different protein kinase selectivities. | ||
| + | </StructureSection> | ||
| + | __NOTOC__ | ||
===3D structures of glycogen synthase kinase 3=== | ===3D structures of glycogen synthase kinase 3=== | ||
| Line 17: | Line 22: | ||
[[3m1s]], [[3pup]] - hGSK-3 β + Ru complex<br /> | [[3m1s]], [[3pup]] - hGSK-3 β + Ru complex<br /> | ||
[[3zdi]] - hGSK-3 β + Axin peptide + inhibitor | [[3zdi]] - hGSK-3 β + Axin peptide + inhibitor | ||
| - | + | <references/> | |
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Revision as of 12:03, 3 November 2013
| |||||||||||
3D structures of glycogen synthase kinase 3
Updated on 03-November-2013
1i09, 1h8f – hGSK-3 β – human
1gng - hGSK-3 β + Frattide peptide
1o9u - hGSK-3 β + Axin peptide
1q4l, 1uv5, 1q5k, 1r0e, 2o5k, 2ow3, 3du8, 3f7z, 3f88, 3i4b, 3gb2, 3l1s, 3q3b, 3zrk, 3zrl, 3zrm, 3sd0, 4dit, 4afj, 4acc, 4acd, 4acg, 4ach, 3say - hGSK-3 β + inhibitor
1pyx, 1j1b - hGSK-3 β + AMPPNP
1j1c - hGSK-3 β + ADP
1q3d, 1q3w, 1q41 - hGSK-3 β + ATP-mimetic inhibitor
2jld - hGSK-3 β + Ru complex + peptide
3m1s, 3pup - hGSK-3 β + Ru complex
3zdi - hGSK-3 β + Axin peptide + inhibitor
- ↑ Atilla-Gokcumen GE, Di Costanzo L, Meggers E. Structure of anticancer ruthenium half-sandwich complex bound to glycogen synthase kinase 3beta. J Biol Inorg Chem. 2010 Sep 7. PMID:20821241 doi:10.1007/s00775-010-0699-x