We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Ku protein
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
=== Ku Ring === | === Ku Ring === | ||
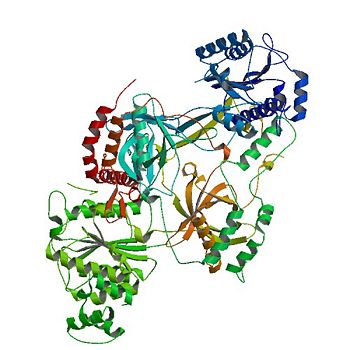
| - | The <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> is composed of a broad base of beta barrels that cradle the DNA, and a narrow bridge that serves to protect the double strand break from base pairing with other DNA base pairs and degradation <ref name="Walker"/>. There is little interaction between the ring and the backbone or base pairs of DNA; instead, the ring associates with DNA by the cradle fitting into the major grooves of the helix <ref name="Walker"/>. The positive electrostatic charge caused by polarization of the ring also allows the negatively charged backbone of DNA to be guided into the correct position <ref name="Walker"/> | + | The <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> is composed of a broad base of beta barrels that cradle the DNA, and a narrow bridge that serves to protect the double strand break from base pairing with other DNA base pairs and degradation <ref name="Walker"/>. There is little interaction between the ring and the backbone or base pairs of DNA; instead, the ring associates with DNA by the cradle fitting into the major grooves of the helix <ref name="Walker"/>. The positive electrostatic charge caused by polarization of the ring also allows the negatively charged backbone of DNA to be guided into the correct position <ref name="Walker"/> [[NEED SCENE OF POS CHARGE OR POLARIZATION]]. The Ku protein also has a high affinity to DNA due to its form being preset for the helix. As a result of the asymmetric ring, there is a strong preference (Kd value of 1.5 to 4 X 10^-10 M<ref name="Walker"/>) for the <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> to slide onto the ends of DNA <ref name="1"/>. In addition, other asymmetric features, such as a abundance of Asp residues on the N terminus of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> [[NEED SCENE OF ASP ON N-TERMINUS OR MAYBE JUST ASP IN GENERAL]], prevent the Ku protein from sliding further on the DNA helix. While wrapping over the entire helix, the <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> is thin over the bridge, allowing ligases and polymerases to efficiently interact in [[non-homologous end joining (NHEJ)]]. <ref name="Walker"/> |
== Domains == | == Domains == | ||
<scene name='56/567269/Ku70_subunit/3'>Ku70/80 subunits</scene> | <scene name='56/567269/Ku70_subunit/3'>Ku70/80 subunits</scene> | ||
| - | Consisting of three domains (<scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene>, <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene>, <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene>), the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> dimerizes with the <scene name='56/567269/Ku80_subunit/3'>Ku80 subunit</scene> to form the protein. | + | Consisting of three domains (<scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene>, <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene>, <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene>), the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> dimerizes with the <scene name='56/567269/Ku80_subunit/3'>Ku80 subunit</scene> to form the protein.<ref name="Walker"/> |
| + | Unlike other DNA binding proteins, the Ku protein is asymmetrical from the differences between the Ku70 and Ku80 subunits. | ||
| + | This asymmetry leads to different favorable locations for DNA based on major and minor grooves.<ref name="Walker"/> | ||
| + | The <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> is angled closer to <scene name='56/567269/Bound_dna/3'>DNA</scene> at the double strand break, providing protectiion and interaction with its domains.<ref name="source2"> PMID: 19715578</ref> | ||
| + | In contrast, the <scene name='56/567269/Ku80_subunit/3'>Ku80 subunit</scene> associates with <scene name='56/567269/Bound_dna/3'>DNA</scene> away from the free end.<ref name="Walker"/> Once a homodimer, the protein has diverged into two domains that are now 15% similar in residues. <ref name="source3"> PMID: 9663392</ref> | ||
=== α/β-Domain === | === α/β-Domain === | ||
| - | Contained inside the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> is a [[Rossman fold]] at the N terminus that is used to bind nucleotides in <scene name='56/567269/Bound_dna/3'>DNA</scene> <ref | + | Contained inside the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> is a [[Rossman fold]] at the N terminus that is used to bind nucleotides in <scene name='56/567269/Bound_dna/3'>DNA</scene>.<ref name="Walker"/> |
| + | In terms of protein structure, the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> contributes little to the dimer interface between the subunits. | ||
| + | The C terminus of the domain can be bound to other repair molecules, using <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> as a scaffold.<ref name="Walker"/> | ||
=== β-barrel === | === β-barrel === | ||
| - | The <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> is the main source of interactions of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> itself and <scene name='56/567269/Bound_dna/3'>DNA helix</scene>, with each <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> being composed of seven β strands with the majority in antiparallel arrangement <ref | + | The <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> is the main source of interactions of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> itself and <scene name='56/567269/Bound_dna/3'>DNA helix</scene>, with each <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> being composed of seven β strands with the majority in antiparallel arrangement.<ref name="Walker"/> |
| + | The quantity of the strands lends the structures to be symmetrical. Both <scene name='56/567269/Ku70_dimer/4'>β-barrels</scene> in the dimer form the base of the cradle by fitting in the grooves of <scene name='56/567269/Bound_dna/3'>DNA</scene>. | ||
=== C-terminal arm === | === C-terminal arm === | ||
| - | The <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> is an α-helical domain that associates with the β-barrel of the opposite subunit, with the arm stretching across the <scene name='56/567269/Bound_dna/3'>DNA helix</scene> <ref | + | The <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> is an α-helical domain that associates with the β-barrel of the opposite subunit, with the arm stretching across the <scene name='56/567269/Bound_dna/3'>DNA helix</scene>.<ref name="Walker"/> |
| + | As a result, the <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> strengthens the cradle composed of the two β-barrels. | ||
=== DNA binding ring === | === DNA binding ring === | ||
| - | The <scene name='56/567269/Ku70_dimer/6'>DNA binding ring</scene> on the open end of DNA is associated with the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene>. By binding <scene name='56/567269/Bound_dna/3'>DNA</scene>, Ku realigns the the strands and protects the molecule from degradation and unwanted bonds while NHEJ occurs <ref | + | The <scene name='56/567269/Ku70_dimer/6'>DNA binding ring</scene> on the open end of DNA is associated with the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene>. |
| + | By binding <scene name='56/567269/Bound_dna/3'>DNA</scene>, Ku realigns the the strands and protects the molecule from degradation and unwanted bonds while NHEJ occurs.<ref name="Walker"/> | ||
| + | The regulation of the DNA binding ring of Ku is still under research, with data supporting oxidative stress and redox reactions decreasing the association of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> with <scene name='56/567269/Bound_dna/3'>DNA</scene> through alterations in cysteine residues on the <scene name='56/567269/Ku70_subunit/3'>Ku70 subunit</scene> [[NEED SCENE OF CYSTEINES]]. <ref name="source3"/> <ref name="source4"> PMID: 14585978</ref> | ||
| Line 45: | Line 55: | ||
== Function == | == Function == | ||
| - | The <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> serves to assist in [[non-homologous end joining (NHEJ)]], and also in telomere synthesis and protection. These functions are separate interactions based on key residues that are being identified through current research. Recent research also links the Ku protein with heterochromatin formation through interaction with [[Rif]] and [[Sir proteins]] | + | The <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> serves to assist in [[non-homologous end joining (NHEJ)]], and also in telomere synthesis and protection. These functions are separate interactions based on key residues that are being identified through current research. Recent research also links the Ku protein with heterochromatin formation through interaction with [[Rif]] and [[Sir proteins]]. <ref name="source3"/><ref name="source4"/> |
| + | |||
| - | <ref> PMID: 19715578</ref> | ||
| - | <ref> PMID: 9663392</ref> | ||
| - | <ref> PMID: 14585978</ref> | ||
</StructureSection> | </StructureSection> | ||
Revision as of 20:37, 4 November 2013
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Bennett SM, Neher TM, Shatilla A, Turchi JJ. Molecular analysis of Ku redox regulation. BMC Mol Biol. 2009 Aug 28;10:86. doi: 10.1186/1471-2199-10-86. PMID:19715578 doi:http://dx.doi.org/10.1186/1471-2199-10-86
- ↑ 3.0 3.1 3.2 Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998 Jul 2;8(14):831-4. PMID:9663392
- ↑ 4.0 4.1 Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol. 2003 Nov;23(22):8202-15. PMID:14585978