We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Ku protein
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
=== Ku Ring === | === Ku Ring === | ||
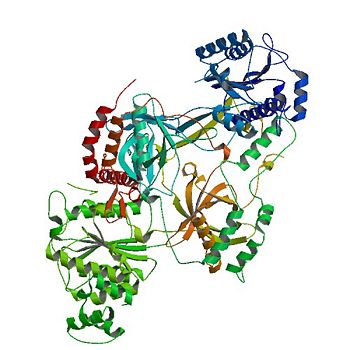
| - | The <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> is composed of a broad base of beta barrels that cradle the DNA, and a narrow bridge that serves to protect the double strand break from base pairing with other DNA base pairs and degradation <ref name="Walker"/>. There is little interaction between the ring and the backbone or base pairs of DNA; instead, the ring associates with DNA by the cradle fitting into the major grooves of the helix <ref name="Walker"/>. The positive electrostatic charge caused by polarization of the ring also allows the negatively charged backbone of DNA to be guided into the correct position <ref name="Walker"/> [[NEED SCENE OF POS CHARGE OR POLARIZATION]]. The Ku protein also has a high affinity to DNA due to its form being preset for the helix. As a result of the asymmetric ring, there is a strong preference (Kd value of 1.5 to 4 X 10^-10 M<ref name="Walker"/>) for the <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> to slide onto the ends of DNA <ref name=" | + | The <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> is composed of a broad base of beta barrels that cradle the DNA, and a narrow bridge that serves to protect the double strand break from base pairing with other DNA base pairs and degradation <ref name="Walker"/>. There is little interaction between the ring and the backbone or base pairs of DNA; instead, the ring associates with DNA by the cradle fitting into the major grooves of the helix <ref name="Walker"/>. The positive electrostatic charge caused by polarization of the ring also allows the negatively charged backbone of DNA to be guided into the correct position <ref name="Walker"/> [[NEED SCENE OF POS CHARGE OR POLARIZATION]]. The Ku protein also has a high affinity to DNA due to its form being preset for the helix. As a result of the asymmetric ring, there is a strong preference (Kd value of 1.5 to 4 X 10^-10 M<ref name="Walker"/>) for the <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> to slide onto the ends of DNA <ref name="Walker"/>. In addition, other asymmetric features, such as a abundance of Asp residues on the N terminus of the <scene name='56/567269/Ku_heterodimer/3'>Ku heterodimer</scene> [[NEED SCENE OF ASP ON N-TERMINUS OR MAYBE JUST ASP IN GENERAL]], prevent the Ku protein from sliding further on the DNA helix. While wrapping over the entire helix, the <scene name='56/567269/Ku_ring/1'>Ku Ring</scene> is thin over the bridge, allowing ligases and polymerases to efficiently interact in [[non-homologous end joining (NHEJ)]]. <ref name="Walker"/> |
== Domains == | == Domains == | ||
Revision as of 20:38, 4 November 2013
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Bennett SM, Neher TM, Shatilla A, Turchi JJ. Molecular analysis of Ku redox regulation. BMC Mol Biol. 2009 Aug 28;10:86. doi: 10.1186/1471-2199-10-86. PMID:19715578 doi:http://dx.doi.org/10.1186/1471-2199-10-86
- ↑ 3.0 3.1 3.2 Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998 Jul 2;8(14):831-4. PMID:9663392
- ↑ 4.0 4.1 Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol. 2003 Nov;23(22):8202-15. PMID:14585978