We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Ku protein
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
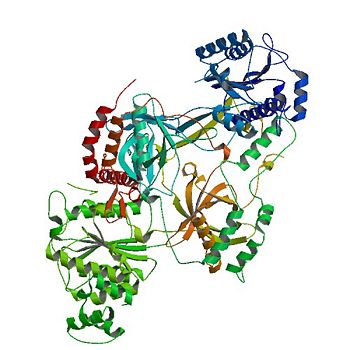
=== α/β-Domain === | === α/β-Domain === | ||
| - | Contained inside the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> is a [[Rossman fold]] at the N terminus that is used to bind nucleotides in | + | Contained inside the <scene name='56/567269/Ku70_dimer/2'>α/β-Domain</scene> is a [[Rossman fold]] at the N terminus that is used to bind nucleotides in DNA.<ref name="Walker"/> |
| - | In terms of protein structure, the | + | In terms of protein structure, the α/β-Domain contributes little to the dimer interface between the subunits. |
| - | The C terminus of the domain can be bound to other repair molecules, using | + | The C terminus of the domain can be bound to other repair molecules, using the α/β-Domain as a scaffold.<ref name="Walker"/> |
=== β-barrel === | === β-barrel === | ||
| - | The <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> is the main source of interactions of the | + | The <scene name='56/567269/Ku70_dimer/4'>β-barrel</scene> is the main source of interactions of the Ku heterodimer itself and DNA helix, with each β-barrel being composed of seven β strands with the majority in antiparallel arrangement.<ref name="Walker"/> |
| - | The quantity of the strands lends the structures to be symmetrical. Both | + | The quantity of the strands lends the structures to be symmetrical. Both β-barrel in the dimer form the base of the cradle by fitting in the grooves of DNA. |
=== C-terminal arm === | === C-terminal arm === | ||
| - | The <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> is an α-helical domain that associates with the β-barrel of the opposite subunit, with the arm stretching across the | + | The <scene name='56/567269/Ku70_dimer/7'>C-terminal arm</scene> is an α-helical domain that associates with the β-barrel of the opposite subunit, with the arm stretching across the DNA helix.<ref name="Walker"/> |
| - | As a result, the | + | As a result, the C-terminal arm strengthens the cradle composed of the two β-barrels. |
Revision as of 21:51, 4 November 2013
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001 Aug 9;412(6847):607-14. PMID:11493912 doi:10.1038/35088000
- ↑ Bennett SM, Neher TM, Shatilla A, Turchi JJ. Molecular analysis of Ku redox regulation. BMC Mol Biol. 2009 Aug 28;10:86. doi: 10.1186/1471-2199-10-86. PMID:19715578 doi:http://dx.doi.org/10.1186/1471-2199-10-86
- ↑ 3.0 3.1 3.2 Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998 Jul 2;8(14):831-4. PMID:9663392
- ↑ 4.0 4.1 Bertuch AA, Lundblad V. The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol. 2003 Nov;23(22):8202-15. PMID:14585978