User:Andrew Wills/Sandbox 1
From Proteopedia
| Line 6: | Line 6: | ||
<nowiki><nowiki>Insert non-formatted text here</nowiki></nowiki> | <nowiki><nowiki>Insert non-formatted text here</nowiki></nowiki> | ||
==General Function== | ==General Function== | ||
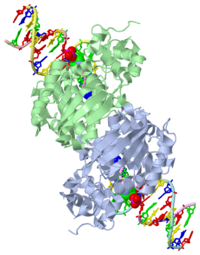
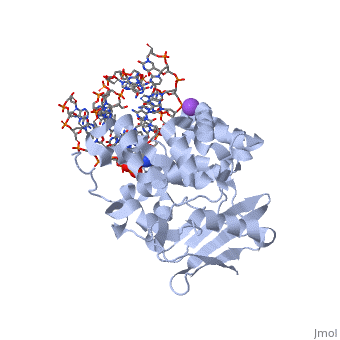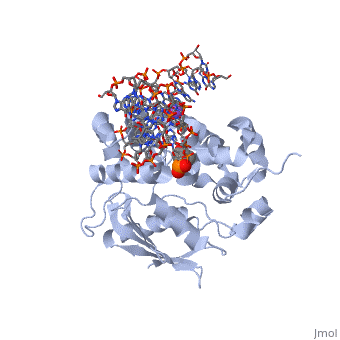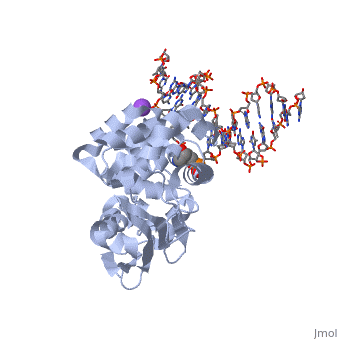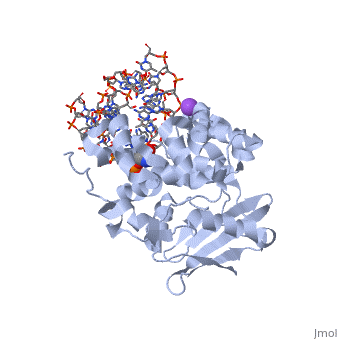
| - | [[1diz]] Escherichia coli 3 methyladenine DNA glycosylase II (<scene name='56/566536/Alka_only/2'>AlkA</scene>) is a DNA repair enzyme that initiates base excision repair for the removal of alkylated bases. Aflatoxin B1 is an example of a toxin that attacks | + | [[1diz]] Escherichia coli 3 methyladenine DNA glycosylase II (<scene name='56/566536/Alka_only/2'>AlkA</scene>) is a DNA repair enzyme that initiates base excision repair for the removal of alkylated bases. Aflatoxin B1 is an example of a toxin that attacks guanosine at its N-7 atom to form alkylated bases<ref name="Berg">Berg, Jeremy, Tymoczko John, and Lubert Stryer. Biochemistry. 6th. New York: W.H. Freeman and Company, 2007. 806-808. Print. </ref> that, which prevent regulatory proteins from binding to DNA and blocks replicative polymerases.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> AlkA initiates base excision repair by first locating and binding to the alkylated DNA. It then flips the affected base out of the DNA double helix and into the active site of the enzyme. Once in the active site, AlkA hydrolyzes the glycosidic bond to release the damaged base and leave the sugar phosphate backbone intact. This creates the AP site that is either devoid of a purine or pyridine. The AP site signals to other base excision repair enzymes to insert an undamaged nucleotide based on the undamaged complementary strand and seal the DNA. |
<StructureSection load='1diz' size='500' side='right' caption='STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA (PDB entry [[1diz]])' scene=''> | <StructureSection load='1diz' size='500' side='right' caption='STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA (PDB entry [[1diz]])' scene=''> | ||
Revision as of 03:40, 6 November 2013
| |||||||||
| 1diz, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Non-Standard Residues: | |||||||||
| Activity: | DNA-3-methyladenine glycosylase II, with EC number 3.2.2.21 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA
<nowiki>Insert non-formatted text here</nowiki>
General Function
1diz Escherichia coli 3 methyladenine DNA glycosylase II () is a DNA repair enzyme that initiates base excision repair for the removal of alkylated bases. Aflatoxin B1 is an example of a toxin that attacks guanosine at its N-7 atom to form alkylated bases[1] that, which prevent regulatory proteins from binding to DNA and blocks replicative polymerases.[2] AlkA initiates base excision repair by first locating and binding to the alkylated DNA. It then flips the affected base out of the DNA double helix and into the active site of the enzyme. Once in the active site, AlkA hydrolyzes the glycosidic bond to release the damaged base and leave the sugar phosphate backbone intact. This creates the AP site that is either devoid of a purine or pyridine. The AP site signals to other base excision repair enzymes to insert an undamaged nucleotide based on the undamaged complementary strand and seal the DNA.
| |||||||||||
See Also
References
- ↑ Berg, Jeremy, Tymoczko John, and Lubert Stryer. Biochemistry. 6th. New York: W.H. Freeman and Company, 2007. 806-808. Print.
- ↑ 2.0 2.1 Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.