This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Andrew Wills/Sandbox 1
From Proteopedia
| Line 22: | Line 22: | ||
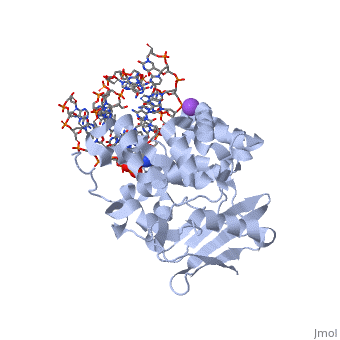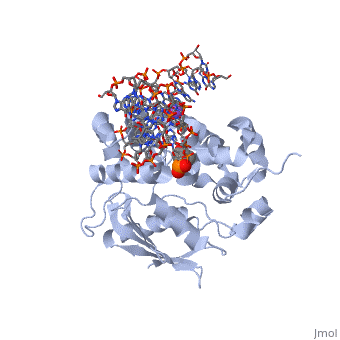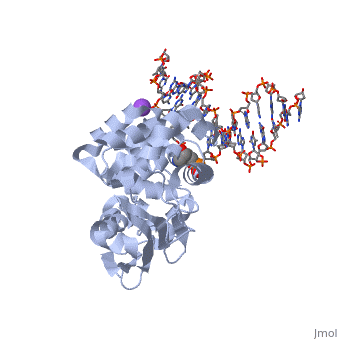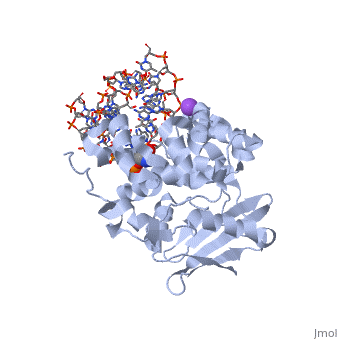
The AlkA protein stabilizes DNA by polar and nonpolar interactions with the DNA backbone. There are few positively charged residues on the AlkA’s binding surface that could interact with the DNA backbone, which is why the majority of polar interactions involve the DNA strand containing the alkylated base. | The AlkA protein stabilizes DNA by polar and nonpolar interactions with the DNA backbone. There are few positively charged residues on the AlkA’s binding surface that could interact with the DNA backbone, which is why the majority of polar interactions involve the DNA strand containing the alkylated base. | ||
| - | DNA is held in place by interactions with the HhH segment, which acts as a platform to create the 66 degree bend in DNA mentioned earlier. The bending of the DNA relieves tensional strain created by the widening of the minor groove for base flipping.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> Amide nitrogens on residues 214, 216, and 219 <scene name='56/566536/Hydrogen_bonding/1'>hydrogen bond</scene> with DNA’s phosphodiester backbone, while sodium metal ion | + | DNA is held in place by interactions with the HhH segment, which acts as a platform to create the 66 degree bend in DNA mentioned earlier. The bending of the DNA relieves tensional strain created by the widening of the minor groove for base flipping.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> Amide nitrogens on residues 214, 216, and 219 <scene name='56/566536/Hydrogen_bonding/1'>hydrogen bond</scene> with DNA’s phosphodiester backbone, while residues 210, 212, and 215 <scene name='56/566536/Coordinate_sodium_ion/1'>coordinate</scene> the sodium metal ion that interacts with DNA further stabilize it in the molecule.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> |
In order to keep the DNA strand bound to the protein, AlkA depends on van der Waals interactions on the minor groove of DNA. The van der Waals interactions play a major role in AlkA’s preference for double stranded DNA.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> An example of this may be seen with the Pro175 wedged into the minor groove of DNA and anchoring it by van der Waals interctions.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> | In order to keep the DNA strand bound to the protein, AlkA depends on van der Waals interactions on the minor groove of DNA. The van der Waals interactions play a major role in AlkA’s preference for double stranded DNA.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> An example of this may be seen with the Pro175 wedged into the minor groove of DNA and anchoring it by van der Waals interctions.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> | ||
Revision as of 04:29, 7 November 2013
| |||||||||
| 1diz, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Non-Standard Residues: | |||||||||
| Activity: | DNA-3-methyladenine glycosylase II, with EC number 3.2.2.21 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
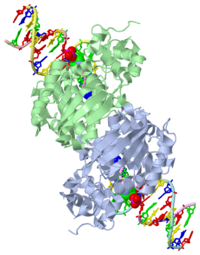
CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA
General Function
1diz Escherichia coli 3 methyladenine DNA glycosylase II (AlkA) is a DNA repair enzyme that initiates base excision repair for the removal of alkylated bases. There are many environmental toxins and cellular agents that may alkylate DNA bases. One example is Aflatoxin, a compound that is converted into a reactive epoxide by cytochrome P450 and attacks guanosine at its N-7 atom to form an alkylated base.[1] The alkylated DNA bases prevent regulatory proteins from binding to DNA and blocks replicative polymerases to stop DNA synthesis.[2] AlkA initiates base excision repair by first locating and binding to the alkylated DNA. It has broad substrate specificity and is able to remove a variety of different alkylated bases. AlkA then flips the affected base out of the DNA double helix and into the active site of the enzyme. Once in the active site, AlkA hydrolyzes the glycosidic bond to release the damaged base and leave the sugar phosphate backbone intact. This creates the AP site that is either devoid of a purine or pyridine. The AP site signals to other base excision repair enzymes to insert an undamaged nucleotide based on the undamaged complementary strand and seal the DNA.[1]
| |||||||||||
See Also
References
- ↑ 1.0 1.1 Berg, Jeremy, Tymoczko John, and Lubert Stryer. Biochemistry. 6th. New York: W.H. Freeman and Company, 2007. 806-808. Print.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.
- ↑ 4.0 4.1 4.2 4.3 Moe, E, D.R. Hall, et al. "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans." Biological Crystallography. 68.6 (2012): 703-712. Print.