Ornithine decarboxylase
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
[[Image:7odc.png|left|200px]] | [[Image:7odc.png|left|200px]] | ||
| - | ===CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION ([[7odc]])=== | + | ===CRYSTAL STRUCTURE ORNITHINE DECARBOXYLASE FROM MOUSE, TRUNCATED 37 RESIDUES FROM THE C-TERMINUS, TO 1.6 ANGSTROM RESOLUTION ([[7odc]]) <ref>PMID:10378276</ref><ref>PMID:18369191</ref>=== |
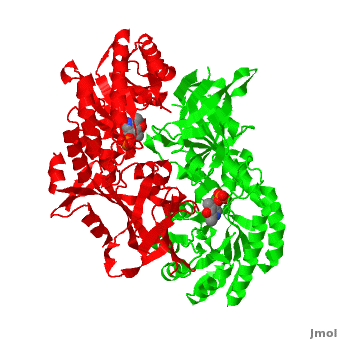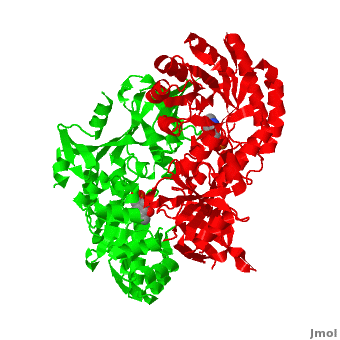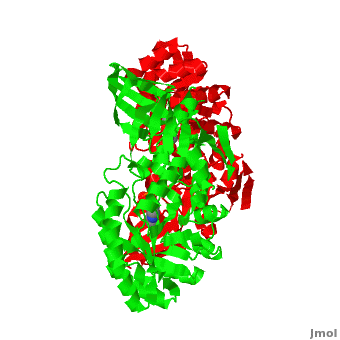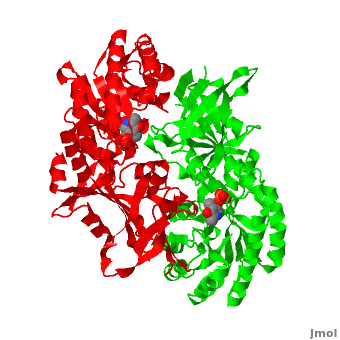
Although, [[7odc]] is a 1 chain structure, the biological relevant molecule for [[7odc]] can be assembled from the contents of the deposited coordinates by the application of crystallographic symmetry operations to give a dimer. It can be [http://www.ebi.ac.uk/pdbe/pqs/macmol/7odc.mmol downloaded]. A sequence alignment and structural comparison of mouse [http://en.wikipedia.org/wiki/Ornithine_decarboxylase ornithine decarboxylase] (mODC) to mouse [[Antizyme Inhibitor]] (AzI, [[3btn]]) show high sequence identity (~50%) and structural similarity between mODC and AzI monomers (RMSD value is 1.6 Å). The <scene name='Antizyme_Inhibitor/Azi_odc/10'>structural comparison</scene> of mODC (<font color='red'><b>red</b></font> and <font color='lime'><b>lime</b></font>) to mouse AzI crystallographic dimer (mAzI, <font color='cyan'><b>cyan</b></font> and <font color='blueviolet'><b>blueviolet</b></font>) is shown. Superposition of the <scene name='Antizyme_Inhibitor/Azi_odc/11'>interface</scene> of mAzI and mODC showing the inter-subunit variable loops (AzI residues 355–362 and 387–401). <font color='black'><b>AzI loops</b></font> are in <font color='black'><b>black</b></font>, and <font color='black'><b>ODC loops</b></font> are in <font color='black'><b>yellow</b></font>. | Although, [[7odc]] is a 1 chain structure, the biological relevant molecule for [[7odc]] can be assembled from the contents of the deposited coordinates by the application of crystallographic symmetry operations to give a dimer. It can be [http://www.ebi.ac.uk/pdbe/pqs/macmol/7odc.mmol downloaded]. A sequence alignment and structural comparison of mouse [http://en.wikipedia.org/wiki/Ornithine_decarboxylase ornithine decarboxylase] (mODC) to mouse [[Antizyme Inhibitor]] (AzI, [[3btn]]) show high sequence identity (~50%) and structural similarity between mODC and AzI monomers (RMSD value is 1.6 Å). The <scene name='Antizyme_Inhibitor/Azi_odc/10'>structural comparison</scene> of mODC (<font color='red'><b>red</b></font> and <font color='lime'><b>lime</b></font>) to mouse AzI crystallographic dimer (mAzI, <font color='cyan'><b>cyan</b></font> and <font color='blueviolet'><b>blueviolet</b></font>) is shown. Superposition of the <scene name='Antizyme_Inhibitor/Azi_odc/11'>interface</scene> of mAzI and mODC showing the inter-subunit variable loops (AzI residues 355–362 and 387–401). <font color='black'><b>AzI loops</b></font> are in <font color='black'><b>black</b></font>, and <font color='black'><b>ODC loops</b></font> are in <font color='black'><b>yellow</b></font>. | ||
| Line 35: | Line 35: | ||
==References== | ==References== | ||
<ref group="xtra">PMID:10378276</ref> <ref group="xtra">PMID:18369191</ref><references group="xtra"/> | <ref group="xtra">PMID:10378276</ref> <ref group="xtra">PMID:18369191</ref><references group="xtra"/> | ||
| + | <references/> | ||
[[Category: Mus musculus]] | [[Category: Mus musculus]] | ||
[[Category: Ornithine decarboxylase]] | [[Category: Ornithine decarboxylase]] | ||
Revision as of 09:11, 13 November 2013
| |||||||||||
3D structures of ornithine decarboxylase
1ord – LaODC + PLP – Lactobacillus
7odc - ODC (mutant) + PLP – mouse
1qu4 - TbODC + PLP – Trypanosoma brucei
1d7k - hODC + PLP – human
2on3 - hODC + aminoxy-aminopropane
2tod - TbODC + PLP + difluoromewthylornithine
1f3t - TbODC + PLP + putrescine
1njj - TbODC + geneticin + ornithine
1szr - TbODC + PLP + ornithine derivative + PLP derivative
1c4k - LaODC (mutant) + PLP + GTP
2oo0 - hODC + aminoxy-aminopropane + PLP + pentane-diamine
Lysine/Ornithine decarboxylase
2plj - VvLODC + putrescine – Vibrio vulnificus
2plk - VvLODC + cadaverine
References
- Kern AD, Oliveira MA, Coffino P, Hackert ML. Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure. 1999 May;7(5):567-81. PMID:10378276
- Albeck S, Dym O, Unger T, Snapir Z, Bercovich Z, Kahana C. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 2008 May;17(5):793-802. Epub 2008 Mar 27. PMID:18369191 doi:10.1110/ps.073427208
- ↑ Kern AD, Oliveira MA, Coffino P, Hackert ML. Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases. Structure. 1999 May;7(5):567-81. PMID:10378276
- ↑ Albeck S, Dym O, Unger T, Snapir Z, Bercovich Z, Kahana C. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 2008 May;17(5):793-802. Epub 2008 Mar 27. PMID:18369191 doi:10.1110/ps.073427208
- ↑ Sanchita, Chauhan R, Soni G, Sudhamalla B, Sharma A. Docking and molecular dynamics studies of peptide inhibitors of ornithine decarboxylase: a rate-limiting enzyme for the metabolism of Fusarium solani. J Biomol Struct Dyn. 2012 Sep 13. PMID:22970930 doi:10.1080/07391102.2012.718526