We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Transfer RNA (tRNA)
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
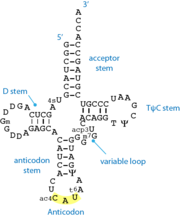
The structure of most tRNA is composed of <scene name='TRNA/Fullview_cartoon/1'>four helical stems</scene> arranged in a cloverleaf structure with four helical stems and an central four-way junction. The <scene name='TRNA/Acceptor_cartoon/1'>acceptor stem</scene> comprises the 5' and 3' ends of the molecule, the latter having an extension of four unpaired nucleotides, with a conserved terminal -CCA sequence at the 3' end. The anticodon stem, at the other end of the molecule is closed by the <scene name='TRNA/Anticodon_cartoon/2'>anticodon</scene> loop. The TΨC-stem and loop and the D-stem and loop form the core of the molecule. | The structure of most tRNA is composed of <scene name='TRNA/Fullview_cartoon/1'>four helical stems</scene> arranged in a cloverleaf structure with four helical stems and an central four-way junction. The <scene name='TRNA/Acceptor_cartoon/1'>acceptor stem</scene> comprises the 5' and 3' ends of the molecule, the latter having an extension of four unpaired nucleotides, with a conserved terminal -CCA sequence at the 3' end. The anticodon stem, at the other end of the molecule is closed by the <scene name='TRNA/Anticodon_cartoon/2'>anticodon</scene> loop. The TΨC-stem and loop and the D-stem and loop form the core of the molecule. | ||
| - | The overall shape of the molecule is that of a capital "L" letter. The two arms of the "L" are formed by the stacking of the acceptor and TΨC-stem on one side, and of the anticodon and D-stem on the other side. <scene name='TRNA/Tertiary_interactions/1'>Tertiary interactions</scene> between the TΨC- and D-loop form the corner of the L-shape and stabilise the structure. | + | The overall shape of the molecule is that of a capital "L" letter. The two arms of the "L" are formed by the stacking of the acceptor and TΨC-stem on one side, and of the anticodon and D-stem on the other side. <scene name='TRNA/Tertiary_interactions/1'>Tertiary interactions</scene> between the TΨC- and D-loop form the corner of the L-shape and stabilise the structure. Non-Watson-Crick <scene name='43/433638/H_bonding/1'>hydrogen bonding</scene> is important in this core. |
In addition, tRNA have a variable loop located in between the acceptor and D-stems. This variable loop can be quite small, but for some tRNA such as the serine or leucine-specific tRNA, it can form an additional helix. | In addition, tRNA have a variable loop located in between the acceptor and D-stems. This variable loop can be quite small, but for some tRNA such as the serine or leucine-specific tRNA, it can form an additional helix. | ||
Revision as of 14:55, 14 November 2013
| |||||||||||
3D Structures of tRNA
Free tRNA
yeast phenylalanine tRNA
human lysine tRNA (primer of HIV1 reverse transcription)
yeast aspartic acid tRNA
E. coli initiatior methionine tRNA
tRNA fragments
Complexes with aminoacyl-tRNA synthetases
Complexes with elongation factors
Complex with RNAse P
Complexes with the ribosome
See Also
References
- ↑ Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010 Jun 22;49(24):4934-44. PMID:20459084 doi:10.1021/bi100408z
Reference for the structure
- Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000 Aug;6(8):1091-105. PMID:10943889
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Wayne Decatur, Michal Harel, Frédéric Dardel, Ann Taylor, Joel L. Sussman, Alexander Berchansky
Categories: Trna | Topic Page | Translation | Modification | RNA | Amino acid