This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox 1k4r
From Proteopedia
| Line 26: | Line 26: | ||
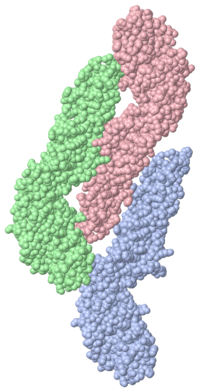
'''Dengue NS3/NS2B Protein'''<StructureSection load='2vbc' size='500' side='right' caption='Structure of protease and helicase (PDB entry [[2vbc]])' scene=''>The NS3 protease is a serine protease that can also function as a RNA helicase and RTPase/NTPase. The enzymatic function of this protease is important for the Dengue virus to replicate. This enzyme of the virus is also a potential target for vaccines and antiviral drugs. | '''Dengue NS3/NS2B Protein'''<StructureSection load='2vbc' size='500' side='right' caption='Structure of protease and helicase (PDB entry [[2vbc]])' scene=''>The NS3 protease is a serine protease that can also function as a RNA helicase and RTPase/NTPase. The enzymatic function of this protease is important for the Dengue virus to replicate. This enzyme of the virus is also a potential target for vaccines and antiviral drugs. | ||
| - | The catalytic triad <scene name='56/565763/Ns2b/3'>(His-51, Asp-75 and Ser-135)</scene>, is found between these two β-barrels, and its activity is dependent on the presence of the <scene name='56/565763/Ns2b/ | + | The catalytic triad <scene name='56/565763/Ns2b/3'>(His-51, Asp-75 and Ser-135)</scene>, is found between these two β-barrels, and its activity is dependent on the presence of the <scene name='56/565763/Ns2b/7'>NS2B</scene>. This cofactor then wraps around the NS3 protease domain and becomes part of the active site. The NS2B cofactor is critical for proteolytic activation of the NS3 protease. The NS3 protease is made up of an extensive network of hydrogen bond and hydrophobic interaction, making it very rigid. NS2B is also important in contributing to substrate binding. This implies that NS2B acts as an enzyme activator as well as being directly involved in substrate binding/interactions.</StructureSection> |
Revision as of 16:01, 14 November 2013
Contents |
Dengue Virus
| |||||||||
| 1k4r, resolution 24.00Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Related: | 1svb | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Structure of Dengue Virus
The first structure of a flavivirus has been determined by using a combination of cryoelectron microscopy and fitting of the known structure of glycoprotein E into the electron density map. The virus core, within a lipid bilayer, has a less-ordered structure than the external, icosahedral scaffold of 90 glycoprotein E dimers. The three E monomers per icosahedral asymmetric unit do not have quasiequivalent symmetric environments. Difference maps indicate the location of the small membrane protein M relative to the overlaying scaffold of E dimers. The structure suggests that flaviviruses, and by analogy also alphaviruses, employ a fusion mechanism in which the distal beta barrels of domain II of the glycoprotein E are inserted into the cellular membrane.
Structure of dengue virus: implications for flavivirus organization, maturation, and fusion., Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH, Cell. 2002 Mar 8;108(5):717-25. PMID:11893341
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
Etiology
Dengue virus is a mosquito borne illness and is a major threat in most of the tropical and sub-tropical countries around the world. There are four related subtypes of the Dengue virus. Dengue is not transmitted directly from person-to-person and symptoms range from a mild fever, to incapacitating high fever, with severe headache, pain behind the eyes, muscle and joint pain, and rash. There is no vaccine or any specific medicine to treat dengue. People who have dengue fever should rest, drink plenty of fluids and reduce the fever using paracetamol or see a doctor.
About this Structure
Dengue Virus Genome carries only one single strand of RNA as its genome. This genome only encodes for ten proteins (three structural and seven "production" proteins. The production proteins are useful in aiding the virus penetrating the host cell and help produce new virus structures. The outer membrane of the protein is constructed of 180 copies of the envelope protein, which this membrane helps the virus attach to the host cell membrane.
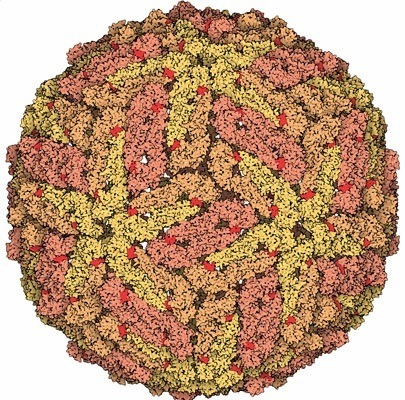 Figure 1: This image represents the entire Dengue genome (complete with 180 copies of the envelope protein)
Figure 1: This image represents the entire Dengue genome (complete with 180 copies of the envelope protein)
| |||||||||||
| |||||||||||
| |||||||||||
Reference
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002 Mar 8;108(5):717-25. PMID:11893341
Categories: Dengue Virus | RCSB PDB Molecule of the Month | Viruses | Baker, T S. | Chipman, P R. | Corver, J. | Jones, C T. | Kuhn, R J. | Lenches, E. | Mukhopadhyay, S. | Pletnev, S V. | Rossmann, M G. | Strauss, E G. | Strauss, J H. | Zhang, W. | Dengue virus | Flaviviridae | Flavivirus | Glycoprotein e from tick-borne encephalitis virus | Icosahedral virus | Virus