We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Andrew Wills/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
==Key Structures== | ==Key Structures== | ||
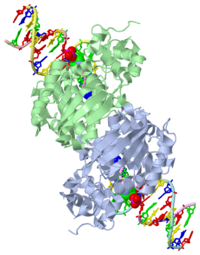
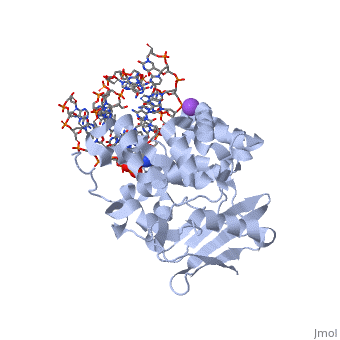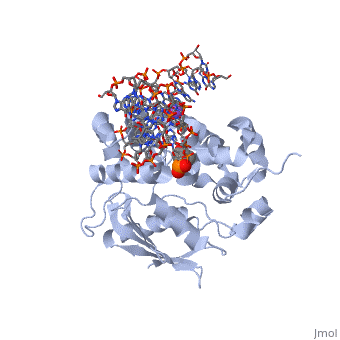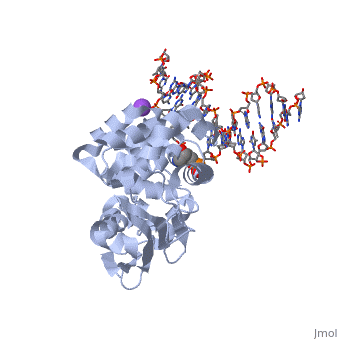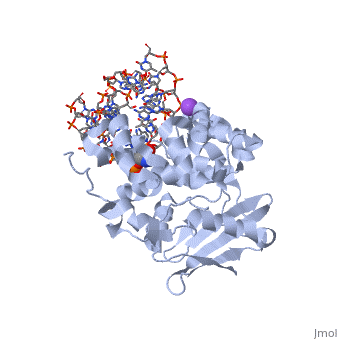
| - | AlkA is composed of three main domains with dimensions of approximately 50 Angstroms, 45 Angstroms, and 25 Angstroms.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The first domain (residues 1-112) is the <scene name='56/566536/N_terminal_domain_and_dna/ | + | AlkA is composed of three main domains with dimensions of approximately 50 Angstroms, 45 Angstroms, and 25 Angstroms.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The first domain (residues 1-112) is the <scene name='56/566536/N_terminal_domain_and_dna/4'>N-Terminal Domain</scene> that is composed of a five stranded antiparallel beta sheet and two alpha helices.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The <scene name='56/566536/Second_domain2/1'>Second Domain</scene> (residues 113-230) contains seven alpha helices that create a hydrophobic core.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The third domain (resudues 231-282) is the <scene name='56/566536/C_terminal_domain/2'>C-Terminal Domain</scene> that contains a bundle of three alpha helices. <ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> The three domains of AlkA create a distinct binding structure that is lined with electron rich aromatic residues to form a hydrophobic cleft. The cleft is found between domains 2 and 3, and forms the binding pocket for the alkylated base. The hydrophobic cleft has the ability to widen between domains 2 and 3 for accepting a variety of alkylated bases.<ref name="Labahn">Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.</ref> |
AlkA is a member of the helix-hairpin-helix (HhH) family of DNA glycosylases, where two compact alpha helical structures are connected by a hairpin loop.<ref name="Moe">Moe, E, D.R. Hall, et al. "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans." Biological Crystallography. 68.6 (2012): 703-712. Print. </ref> In AlkA, the <scene name='56/566536/Helix_hairpin_helix_domain/3'>HhH Domain</scene> is located on the rim of the active site and is composed of residues 202-227.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> The HhH sequence is responsible for binding the damaged DNA by van der Waals interactions, a few hydrogen bonds, and metal ion interactions. | AlkA is a member of the helix-hairpin-helix (HhH) family of DNA glycosylases, where two compact alpha helical structures are connected by a hairpin loop.<ref name="Moe">Moe, E, D.R. Hall, et al. "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans." Biological Crystallography. 68.6 (2012): 703-712. Print. </ref> In AlkA, the <scene name='56/566536/Helix_hairpin_helix_domain/3'>HhH Domain</scene> is located on the rim of the active site and is composed of residues 202-227.<ref name="Hollis">Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print. </ref> The HhH sequence is responsible for binding the damaged DNA by van der Waals interactions, a few hydrogen bonds, and metal ion interactions. | ||
Revision as of 15:30, 19 November 2013
| |||||||||
| 1diz, resolution 2.50Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Non-Standard Residues: | |||||||||
| Activity: | DNA-3-methyladenine glycosylase II, with EC number 3.2.2.21 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA
| |||||||||||
See Also
References
- ↑ 1.0 1.1 Berg, Jeremy, Tymoczko John, and Lubert Stryer. Biochemistry. 6th. New York: W.H. Freeman and Company, 2007. 806-808. Print.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Hollis, Thomas, Yoshitaka Ichikawa, and Tom Ellenberger. "DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA." EMBO Journal. 19.4 (2000): 758-766. Print.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 Labahn, Jorg, Orlando Scharer, et al. "Structural Basis for the Excision Repair of Alkylation-Damaged DNA." Cell. 86.2 (1996): 321-329. Print.
- ↑ 4.0 4.1 4.2 4.3 Moe, E, D.R. Hall, et al. "Structure-function studies of an unusual 3-methyladenine DNA glycosylase II (AlkA) from Deinococcus radiodurans." Biological Crystallography. 68.6 (2012): 703-712. Print.