Sandbox Reserved 776
From Proteopedia
(→Structure) |
(→Structure) |
||
| Line 16: | Line 16: | ||
[[Image:DHP active site.jpg]] | [[Image:DHP active site.jpg]] | ||
| + | |||
| + | |||
| + | [[Image:Dimer interface.gif]] | ||
Revision as of 15:36, 19 November 2013
|
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
Dehaloperoxidase
Dehaloperoxidase (DHP) from the marine annellid Amphitrite ornata is a dimeric enzyme weighing ~16 kDa with a heme prosthetic group and belongs to the globin family. DHP is consistent with the 'globin fold', having eight alpha helices; globins are reversible oxygen binding/transporting proteins (1). Although belonging to the globin superfamily, DHP has a biologically relevant peroxidase activity, the first known globin-peroxidase. It has previously been shown to dehalogenate trihalophenols to their corresponding dihaloquinones. Peroxidases catalyze the reaction of various substrates utilizing hydrogen peroxide as the electron acceptor and [O] insertion to the product originates from water. Reactivity is initiated by use of H2O2 as co-substrate. (2) 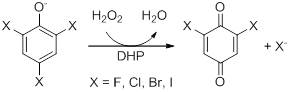
There are two known isoenzymes of DHP, A and B. DHP B has been shown to be more reactive than DHP A, although DHP A has been more extensively studied in the past. The isoenzymes differ at only five residues; I9L, R32K, Y34N, R81S and S91G. (3) More recent work has shown that DHP also reacts through a peroxygenase mechanism by substrate turnover with haloindoles and oxidase activity with indigo formation from 5-bromo-3-oxindole (paper in revision).
Structure
DHP is an enzyme with 127 residues and primarily a dimer in solution which has only been crystallized in its dimeric form. Although DHP belongs to the globin superfamily, it is rich in aspartic and glutamic acids similar to most class III peroxidases. Important for the catalytic activity, DHP has proximal and distal histidines, H170 and H42 respectively. H170 is responsible for the only covalent linkage between heme iron and peptide chain in the fifth coordination site, while H42 is responsible for the stabilization and hydrogen bonding with axial ligands such as water, peroxide and cyanide. The active site also contains Arg38 which helps in the stability of ligands as well as substrates.
