This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Trypsin
From Proteopedia
| Line 39: | Line 39: | ||
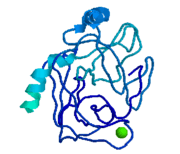
Trypsin, chymotrypsin, and elastase are all digestive enzymes that are produced in the pancreas and catalyze the hydrolysis of peptide bonds. Each of these enzymes has different specificities in regards to the side chains next to the peptide bond. Chymotrypsin prefers a large hydrophobic residue, trypsin is specific for a positively charged residue, and elastase prefers a small neutral residue. Chymotrypsin, trypsin and elastase are all proteins that contain a catalytic mechanism and hydrolyze peptides using the serine protease mechanism. Chymotrypsin and elastase are both homologs of Trypsin since they are 40% alike in structure and composition <ref> Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008. </ref>. In the <scene name='Sandbox_32/Chymotrypsin/2'>Chymotrypsin</scene> structure shown the alpha helices are blue, the beta sheets are green, and the remainder of the protein is red. In the <scene name='Sandbox_32/Elastase/2'>Elastase</scene> structure shown the alpha helices are in red, the beta sheets are yellow, and the remainder of the protein is orange. | Trypsin, chymotrypsin, and elastase are all digestive enzymes that are produced in the pancreas and catalyze the hydrolysis of peptide bonds. Each of these enzymes has different specificities in regards to the side chains next to the peptide bond. Chymotrypsin prefers a large hydrophobic residue, trypsin is specific for a positively charged residue, and elastase prefers a small neutral residue. Chymotrypsin, trypsin and elastase are all proteins that contain a catalytic mechanism and hydrolyze peptides using the serine protease mechanism. Chymotrypsin and elastase are both homologs of Trypsin since they are 40% alike in structure and composition <ref> Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008. </ref>. In the <scene name='Sandbox_32/Chymotrypsin/2'>Chymotrypsin</scene> structure shown the alpha helices are blue, the beta sheets are green, and the remainder of the protein is red. In the <scene name='Sandbox_32/Elastase/2'>Elastase</scene> structure shown the alpha helices are in red, the beta sheets are yellow, and the remainder of the protein is orange. | ||
| + | === The remarkable efficiency of a Pin-II proteinase inhibitor sans two conserved disulfide bonds is due to enhanced flexibility and hydrogen-bond density in the reactive loop <ref>doi 10.1080/07391102.2012.745378</ref> === | ||
| + | |||
| + | Background: Plant proteinase Inhibitors (PIs) are ubiquitous in the plant kingdom and have been extensively studied as plant defense molecules, which inhibit hydrolytic enzymes (''e.g.'' <scene name='Journal:JBSD:39/Cv/12'>trypsin</scene>, <font color='darkmagenta'><b>colored in darkmagenta</b></font>) of the insect gut <ref name="Green">PMID: 17836138</ref>. Among various PI families, Serine PI Pin-II/Pot-II family displays a remarkable structural and functional diversity at the gene and protein level <ref name="Kong">PMID: 18315854</ref>. Wound, herbivory and stress induced up-regulation of these PIs clearly link them to plant defense <ref name="Green">PMID: 17836138</ref>. Previous studies using transgenic systems or in vivo assays have positively correlated the advantage offered by Pin-II PI expression in plants against insect attack <ref name="Johnson">PMID: 2602379</ref> <ref name="Duan">PMID: 9630927</ref>. Precursor proteins of Pin-II PIs consist of 1- to 8- <scene name='Journal:JBSD:39/Cv/4'>inhibitory repeat domains (IRDs)</scene> connected by proteolytic-sensitive linkers, which releases IRD units upon cleavage. <scene name='Journal:JBSD:39/Cv/5'>Each IRD is a peptide of around 50 aa length</scene> (<span style="color:lime;background-color:black;font-weight:bold;">colored in green</span>) with a molecular mass of ~6 KDa. The aa sequence of IRDs shows variations, at the same time the <scene name='Journal:JBSD:39/Cv/6'>8 cysteine residues that form disulfide bridge are conserved</scene> (<span style="color:yellow;background-color:black;font-weight:bold;">colored in yellow</span>) <ref name="Nielsen">PMID: 7578034</ref> <ref name="Scanlon">PMID: 10425681</ref> <ref name="Lee">PMID: 10360353</ref> <ref name="Schirra">PMID: 11178894</ref>. One structural feature of Pin-II IRD is a disordered loop with triple stranded β sheet scaffold. The disordered solvent exposed reactive loop is anchored by the four conserved disulfide bonds (C4-C41, C7-C25, C8-C37 and C14-C50) <ref name="Schirra1">PMID: 16029154</ref> <ref name="Schirra2">PMID: 18991765</ref>. Among the four disulfide bonds, C8-C37 has been found to be very crucial for maintaining active conformation, whereas C4-C41 has an important role in maintaining the flexibility of the reactive loop <ref name="Schirra3">PMID: 19925809</ref>. Thus, any selective loss of disulfide bond is expected to have evolutionary significance leading to functional differentiation of inhibitors <ref name="Li">PMID: 21494600</ref>. | ||
| + | |||
| + | [A] Functionality: To assess the effect of aa variations on activity and structural stability different biochemical studies and 20 ns MD simulations was performed on IRD structures. Inhibition kinetic studies displayed a sigmoidal pattern with increasing concentrations of the inhibitors suggesting reversible and competitive inhibition with tight binding. IRD-9 turned out to be a stronger inhibitor of bovine trypsin (IC50 ~0.0022 mM) than IRD-7 (IC50 ~0.135 mM) and IRD-12 (IC50 ~0.065 mM). | ||
| + | |||
| + | [B] <scene name='Journal:JBSD:39/Cv/16'>Structural Variability</scene>: In accordance with the structure of a typical IRD belonging to Pin-II PI family, the predicted structures of CanPI also have <scene name='Journal:JBSD:39/Cv/7'>three antiparallel β sheets joined by disordered loops containing the reactive site and stabilized by four disulfide bonds</scene>. It was thought that the disulfide bonds act as structural scaffold to hold the reactive site in a relatively rigid conformation and provide thermal and proteolytic stability. A single 3<sub>10</sub>-helix of one turn is also present in the structure, the disordered loop is held by disulfide bond in IRD-7 and -12 whereas by a network of intra molecular hydrogen bonds in IRD-9. <scene name='Journal:JBSD:39/Cv/13'>IRD-7</scene> <span style="color:salmon;background-color:black;font-weight:bold;">(colored in salmon)</span> and <scene name='Journal:JBSD:39/Cv/9'>IRD-12</scene> <span style="color:deeppink;background-color:black;font-weight:bold;">(in deeppink)</span> have 4 disulfide bonds, whereas <scene name='Journal:JBSD:39/Cv/14'>IRD-9</scene> <font color='magenta'><b>(in magenta)</b></font> has only 2 disulfide bonds. Furthermore, post-simulation analysis of the intramolecular hydrogen bonds illustrated that IRD-9 with two disulfide bonds (C7-C25 and C8-C37) less, has a relatively higher density of intra-molecular hydrogen bonds as compared to IRD-7 and -12. These intramolecular hydrogen bonds might be substituting the two lost disulfide bonds of IRD-9 to stabilize the protein structure in the active conformation and might be protecting the molecules from a hydrophobic collapse. The replaced serine residues in the place of two cysteines C7 and C8 in IRD-9 may be contributing to the increased number of hydrogen bonds. | ||
| + | |||
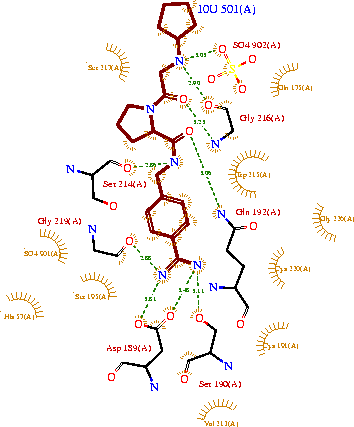
| + | [C] The molecular models of the IRD bound HaTry predicted several atomic interactions with a reactive loop of inhibitors that also explained the contribution of the solvent exposed reactive loop. There are several hydrogen bonds in the <scene name='Journal:JBSD:39/Ird9/3'>IRD-9-HaTry complex</scene>. ARG-39 from <scene name='Journal:JBSD:39/Cv/17'>IRD-12</scene> reactive site formed two hydrogen bonds with the residues of the HaTry active site. In <scene name='Journal:JBSD:39/Ird9/2'>case of IRD-7</scene>, side chain of LYS-39 residue of reactive loop form one hydrogen bond each, with carboxyl oxygen atom of HIS-50. MD simulations provides structural insight into an importance of inter/intra molecular hydrogen bonds and its effect on the interaction between protease and PIs. The results of this analysis were corroborated with previous reports. Post simulation analysis also explained experimentally observed increase in binding affinity, hence activity of IRD-9 towards proteases. See also <ref name="Barrette-Ng">PMID: 12684499</ref> <ref name="Dunse">PMID: 20696921</ref> <ref name="Tamhane">PMID: 19393726</ref> <ref name="Tamhane1">PMID: 15715970</ref>. | ||
</StructureSection> | </StructureSection> | ||
__NOTOC__ | __NOTOC__ | ||
Revision as of 09:57, 20 November 2013
| |||||||||||
3D structures of Trypsin
Updated on 20-November-2013
Cationic trypsin
3nk8, 3nkk, 3mi4, 3mfj, 3iti, 2d8w, 2by5, 2by6, 2by7, 2by8, 2by9, 2bya, 2blv, 2blw, 2a7h, 1s0q, 1uto, 1utp, 1utq, 1utn, 1n6x, 1n6y, 1hj9, 2ptn, 3ptn, 5ptp, 3t25, 3t26, 3t27, 3t28, 3t29, 3unr, 4i8g, 4i8h, 4i8j, 4i8k, 4i8l - bTry1 - bovine
3qk1 – bTry1 (mutant)
1utk, 1utj, 1utl, 1utm, 1hj8 – Try1 – Salmon
1trn – hTry1 – human
3ljj, 3ljo, 3a7t, 3a7v, 3a7w, 3a7x, 3a7y, 3a7z, 3a80, 3a81, 3a82, 3a83, 3a84, 3a85, 3a86, 3a87, 3a88, 3a89, 3a8b, 3a8a, 3a8c, 3a8d, 3m35, 3aas, 3aau, 3aav, 3gy2, 3gy3, 3gy4, 3gy5, 3gy6, 3gy7, 3gy8, 2zq1, 2zq2, 2zhd, 2zfs, 2zft, 2zdk, 2zdl, 2zdm, 2zdn, 2oxs, 2otv, 2g8t, 2g5n, 2g5v, 2ah4, 2fx4, 2fx6, 1yp9, 2ayw, 1y3u, 1y3v, 1y3w, 1y3x, 1y3y, 1tx8, 1tx7, 1s0r, 1rxp, 1o2q, 1o2r, 1o2s, 1o2t, 1o2u, 1o2v, 1o2w, 1o2x, 1o2y, 1o2z, 1o30, 1o31, 1o32, 1o33, 1o34, 1o35, 1o36, 1o37, 1o38, 1o39, 1o3a, 1o3b, 1o3c, 1o3d, 1o3e, 1o3f, 1o3g, 1o3h, 1o3i, 1o3j, 1o3k, 1o3l, 1o3m, 1o3n, 1o3o, 1o3p, 1o2l, 1o2k, 1o2j, 1o2i, 1o2h, 1o2m, 1o2n, 1o2o, 1o2p, 1lqe, 1oyq, 1eb2, 1k1i, 1k1j, 1k1l, 1k1m, 1k1n, 1k1o, 1k1p, 1g36, 1j8a, 1jir, 1g3b, 1g3c, 1g3d, 1g3e, 1g9i, 1f0t, 1f0u, 1c1n, 1c1o, 1c1p, 1c1q, 1c1r, 1c1s, 1c1t, 1c2d, 1c2e, 1c2f, 1c2g, 1c2h, 1c2i, 1c2j, 1c2k, 1c2l, 1c2m, 1qbn, 1qbo, 1qb9, 1qb1, 1qb6, 1qa0, 1qcp, 1ce5, 2bza, 1az8, 1xuf, 1xug, 1bju, 1bjv, 1xuh, 1xui, 1xuj, 1xuk, 1auj, 2tio, 1tio, 1aq7, 3ati, 3atk, 3atl, 3atm, 3rxa, 3rxb, 3rxc, 3rxd, 3rxe, 3rxf, 3rxg, 3rxh, 3rxi, 3rxj, 3rxk, 3rxl, 3rxm, 3rxo, 3rxq, 3rxr, 3rxs, 3rxt, 3rxu, 3rxv - bTry1 + small molecule inhibitor
1v2j, 1v2l, 1v2m, 1v2n, 1v2o, 1v2p, 1v2q, 1v2r, 1v2s, 1v2t, 1v2u, 1v2v, 1v2w, 3plb, 3plk, 3plp, 3pm3, 3pmj, 3pwb, 3pwc, 3pyh, 3q00, 3unq, 3uns, 3uop, 3upe, 3uqo, 3uqv, 3uuz, 3uwi, 3uy9, 3v0x, 3v12, 3v13 - bTry1 (mutant) + small molecule inhibitor
3m7q, 2xtt, 3e8l, 3otj, 3i29, 3d65, 2qyi, 2qn5, 2o9q, 2plx, 2cmy, 2iln, 2uuy, 2j9n, 2g81, 2age, 2agg, 2agi, 2ftl, 2ftm, 2fi3, 2fi4, 2fi5, 1zr0, 1ox1, 1p2i, 1p2j, 1p2k, 1ejm, 1f2s, 3bte, 3btq, 3btd, 3btf, 3btg, 3bth, 3btk, 3btm, 3btt, 3btw, 2btc, 1sbw, 1taw, 1smf, 1ppc, 1ppe, 1pph, 2tld, 1tab, 1tpa, 1c9t, 1ezx, 2f3c, 3rdz – bTry1 + proteinase inhibitor
3ru4 – BTry1 + chymotrypsinogen
4b2b, 4b1t, 4b2a, 4b2c – bTry1 (mutant) + eglin (mutant)
2ra3, 1oph, 3veq - bTry1 (mutant) + proteinase inhibitor
1jrs, 1jrt, 1sfi, 1yyy, 1zzz, 4abi – bTry1 + polypeptide
1c5p, 1c5q, 1c5r, 1c5s, 1c5t, 1c5u, 1c5v, 1ghz, 1gi0, 1gi1, 1gi2, 1gi3, 1gi4, 1gi5, 1gi6, 1gj6, 1mts, 1mtu, 1mtv, 1mtw, 1ql7, 1ql8, 1ql9, 1v2k, 1y59, 1y5a, 1y5b, 1y5u, 3rxp, 4ab8, 4ab9, 4aba, 4abb, 4abd, 4abe, 4abf, 4abg, 4abh, 3vpk – bTry1 + inhibitor
4abj – bTry1 + Try inhibitor 1
4aoq, 4aor - bTry1 + Try inhibitor 3
4j2y - bTry1 + Try inhibitor
2eek – Try1 + inhibitor – Atlantic cod
Cationic trypsinogen
1tgc, 1tgt, 2tga, 2tgt, 1tgb, 1tld, 1tpo - bTryp1
1ntp - β-bTry1 – Neutron diffraction
1d6r, 4tpi, 1tgs, 2tgp, 3tpi, 2tpi, 2ptc - bTryp1 + proteinase inhibitor
1max, 1may, 1btp, 1bty, 1tps, 1tyn, 1tng, 1tnh, 1tni, 1tnj, 1tnk, 1tnl, 1gbt, 1tpp, 3ptb - bTry1 + small molecule inhibitor
1btw, 1btx, 1btz - bTry1 + polypeptide
Anionic trypsin
2zpq, 2zpr, 2zps, 1mbq – Try2 – Chum salmon
1bit, 2tbs - AsTry2 – Atlantic salmon
2sta, 2stb, 1bzx - AsTry2 + proteinase inhibitor
1a0j - AsTry2 + small molecule inhibitor
1ane, 1bra - rTry2]] - rat
1amh, 1dpo, 1anb, 1anc, 1and, 1trm, 2trm - rTry2 (mutant)
3fp6, 3tgi, 1brb, 1brc – rTry2 + proteinase inhibitor
3fp7, 3fp8, 1ykt, 1ylc, 1yld, 1co7, 1k9o, 1slu, 1slv, 1slw, 1slx - rTry2 (mutant) + proteinase inhibitor
1j14, 1j15, 1j16, 1j17 - rTry2 (mutant) + small molecule inhibitor
Anionic trypsinogen
1f5r, 1f7z, 3tgk, 1ezs, 1ezu, 3tgj - rTryp2 (mutant) + proteinase inhibitor
1fy8 - rTryp2 + proteinase inhibitor
Trypsinogen
1tgn – bTryp
2tgd – bTryp + inhibitor
Mesotrypsin
3l33 – hTry3 (mutant) + amyloid β A4
3l3t - hTry3 residues 28-251 (mutant) + amyloid β precursor
2r9p – hTry3 (mutant) + BPTI
Brain trypsin
1h4w – hTry4 + small molecule inhibitor
Neurotrypsin
2k4r, 2k51 – rNTry Kringle domain – NMR
Streptomyces griseus trypsin
3i77, 3i78, 1os8, 1sgt – SGT – Streptomyces griseus
3beu, 2fmj – SGT (mutant)
1oss - SGT (mutant) + small molecule inhibitor
1s81 – pTry – pig
1aks - α-pTry
1ept - ε-pTry
1mct - β-pTry + proteinase inhibitor
3myw, 1yf4, 1z7k, 1tx6, 1v6d, 1uhb, 1h9h, 1h9i, 1eja, 1c9p, 1avw, 1avx, 1ldt, 1tfx, 1an1, 4an7 – pTry + proteinase inhibitor
2a31, 2a32, 1s5s, 1s6f, 1s6h, 1s82, 1s83, 1s84, 1s85, 1fmg, 1fn6, 1fni, 1qqu – pTry + small molecule inhibitor
2vu8 – Try + proteinase inhibitor – mold
2g51, 2g52, 2g55, 1xvo, 1pq5, 1pq7 – FoTry – Fusarium oxysporum
1ppz, 1pqa, 1try - FoTry + small molecule inhibitor
1xvm, 1pq8, 1fn8, 1fy4, 1fy5, 1gdn, 1gdq, 1gdu – FoTry + polypeptide
2f91 – Try-hepatopancreas - Crayfish
References
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Trypsin. 30 October 2010 <http://www.sigmaaldrich.com/life-science/metabolomics/enzyme-explorer/analytical-enzyme/trypsin.html>.
- ↑ Image From: http://chemistry.umeche.maine.edu/MAT500/Peptidase1.html
- ↑ Trypsin. 2010. 30 October 2010 <http://www.worthington-biochem.com/tyr/default.html>
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
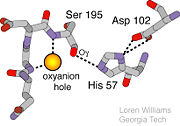
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Image From:

- ↑ Williams, Loren. Georgia Tech. http://www2.chemistry.gatech.edu/~1W26/bcourse_information/6521/protein/serine_protease/triad_1/html.
- ↑ Structural Biochemistry. 10 June 2010. 30 October 2010.<http://en.wikibooks.org/wiki/Structural_Biochemistry/Enzyme/Catalytic_Triad>.
- ↑ Pratt, C.W., Voet, D., Voet, J.G. Fundamentals of Biochemistry - Life at the Molecular Level - Third Edition. Voet, Voet and Pratt, 2008.
- ↑ Joshi RS, Mishra M, Tamhane VA, Ghosh A, Sonavane U, Suresh CG, Joshi R, Gupta VS, Giri AP. The remarkable efficiency of a Pin-II proteinase inhibitor sans two conserved disulfide bonds is due to enhanced flexibility and hydrogen bond density in the reactive site loop. J Biomol Struct Dyn. 2012 Dec 20. PMID:23256852 doi:10.1080/07391102.2012.745378
- ↑ 12.0 12.1 Green TR, Ryan CA. Wound-Induced Proteinase Inhibitor in Plant Leaves: A Possible Defense Mechanism against Insects. Science. 1972 Feb 18;175(4023):776-7. PMID:17836138 doi:10.1126/science.175.4023.776
- ↑ Kong L, Ranganathan S. Tandem duplication, circular permutation, molecular adaptation: how Solanaceae resist pests via inhibitors. BMC Bioinformatics. 2008;9 Suppl 1:S22. PMID:18315854 doi:10.1186/1471-2105-9-S1-S22
- ↑ Johnson R, Narvaez J, An G, Ryan C. Expression of proteinase inhibitors I and II in transgenic tobacco plants: effects on natural defense against Manduca sexta larvae. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9871-5. PMID:2602379
- ↑ Duan X, Li X, Xue Q, Abo-el-Saad M, Xu D, Wu R. Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol. 1996 Apr;14(4):494-8. PMID:9630927 doi:10.1038/nbt0496-494
- ↑ Nielsen KJ, Heath RL, Anderson MA, Craik DJ. Structures of a series of 6-kDa trypsin inhibitors isolated from the stigma of Nicotiana alata. Biochemistry. 1995 Nov 7;34(44):14304-11. PMID:7578034
- ↑ Scanlon MJ, Lee MC, Anderson MA, Craik DJ. Structure of a putative ancestral protein encoded by a single sequence repeat from a multidomain proteinase inhibitor gene from Nicotiana alata. Structure. 1999 Jul 15;7(7):793-802. PMID:10425681
- ↑ Lee MC, Scanlon MJ, Craik DJ, Anderson MA. A novel two-chain proteinase inhibitor generated by circularization of a multidomain precursor protein. Nat Struct Biol. 1999 Jun;6(6):526-30. PMID:10360353 doi:10.1038/9293
- ↑ Schirra HJ, Scanlon MJ, Lee MC, Anderson MA, Craik DJ. The solution structure of C1-T1, a two-domain proteinase inhibitor derived from a circular precursor protein from Nicotiana alata. J Mol Biol. 2001 Feb 9;306(1):69-79. PMID:11178894 doi:10.1006/jmbi.2000.4318
- ↑ Schirra HJ, Craik DJ. Structure and folding of potato type II proteinase inhibitors: circular permutation and intramolecular domain swapping. Protein Pept Lett. 2005 Jul;12(5):421-31. PMID:16029154
- ↑ Schirra HJ, Anderson MA, Craik DJ. Structural refinement of insecticidal plant proteinase inhibitors from Nicotiana alata. Protein Pept Lett. 2008;15(9):903-9. PMID:18991765
- ↑ Schirra HJ, Guarino RF, Anderson MA, Craik DJ. Selective removal of individual disulfide bonds within a potato type II serine proteinase inhibitor from Nicotiana alata reveals differential stabilization of the reactive-site loop. J Mol Biol. 2010 Jan 22;395(3):609-26. Epub 2009 Nov 17. PMID:19925809 doi:10.1016/j.jmb.2009.11.031
- ↑ Li XQ, Zhang T, Donnelly D. Selective loss of cysteine residues and disulphide bonds in a potato proteinase inhibitor II family. PLoS One. 2011 Apr 11;6(4):e18615. PMID:21494600 doi:10.1371/journal.pone.0018615
- ↑ Barrette-Ng IH, Ng KK, Cherney MM, Pearce G, Ryan CA, James MN. Structural basis of inhibition revealed by a 1:2 complex of the two-headed tomato inhibitor-II and subtilisin Carlsberg. J Biol Chem. 2003 Jun 27;278(26):24062-71. Epub 2003 Apr 8. PMID:12684499 doi:10.1074/jbc.M302020200
- ↑ Dunse KM, Kaas Q, Guarino RF, Barton PA, Craik DJ, Anderson MA. Molecular basis for the resistance of an insect chymotrypsin to a potato type II proteinase inhibitor. Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15016-21. Epub 2010 Aug 9. PMID:20696921 doi:10.1073/pnas.1009327107
- ↑ Tamhane VA, Giri AP, Kumar P, Gupta VS. Spatial and temporal expression patterns of diverse Pin-II proteinase inhibitor genes in Capsicum annuum Linn. Gene. 2009 Aug 1;442(1-2):88-98. Epub 2009 Apr 22. PMID:19393726 doi:10.1016/j.gene.2009.04.012
- ↑ Tamhane VA, Chougule NP, Giri AP, Dixit AR, Sainani MN, Gupta VS. In vivo and in vitro effect of Capsicum annum proteinase inhibitors on Helicoverpa armigera gut proteinases. Biochim Biophys Acta. 2005 Mar 11;1722(2):156-67. Epub 2005 Jan 12. PMID:15715970 doi:10.1016/j.bbagen.2004.12.017
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Eran Hodis, Leah Bowlin, David Canner, Glenn Jones, Ben Hallowell, Karl Oberholser, Jaime Prilusky