Single stranded binding protein
From Proteopedia
(→Structure of ''E. coli'' SSB) |
|||
| Line 11: | Line 11: | ||
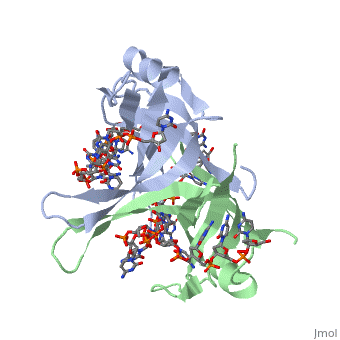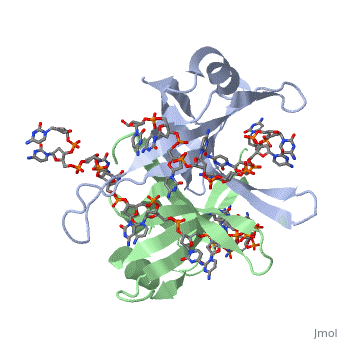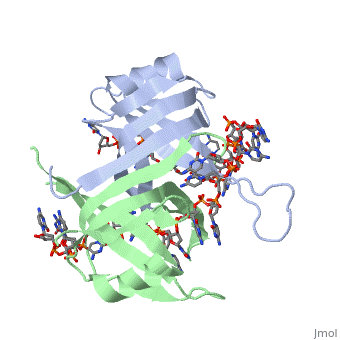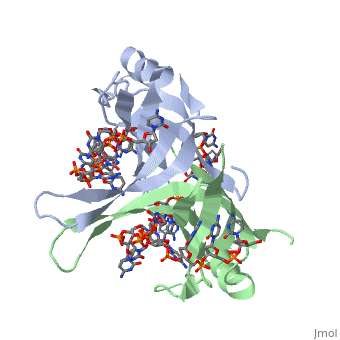
Active ''E. coli'' SSB is made of a homotetramer with extensive DNA binding domains that bind to <scene name='56/566528/Dna/1'>a single strand of DNA</scene><ref>PMID:2087220</ref>. The tetramers consist of <scene name='56/566528/E_coli_ssb_alpha_helices/1'>α-helices</scene>, <scene name='56/566528/Beta_sheets/2'>β-sheets</scene>, and random coils. Each subunit contains an α-helix and several β-sheets. The secondary structure also includes a NH2 terminus, which consists of multiple basic residues, or <scene name='56/566528/Basic_residues/2'>positively charged amino acids</scene>. The DNA-binding domain lies within 115 amino acid residues from this terminus. The COOH terminus includes many negatively charged, or <scene name='56/566528/Acidic_residues/5'>acidic amino acids</scene> <ref>PMID:2087220</ref>. | Active ''E. coli'' SSB is made of a homotetramer with extensive DNA binding domains that bind to <scene name='56/566528/Dna/1'>a single strand of DNA</scene><ref>PMID:2087220</ref>. The tetramers consist of <scene name='56/566528/E_coli_ssb_alpha_helices/1'>α-helices</scene>, <scene name='56/566528/Beta_sheets/2'>β-sheets</scene>, and random coils. Each subunit contains an α-helix and several β-sheets. The secondary structure also includes a NH2 terminus, which consists of multiple basic residues, or <scene name='56/566528/Basic_residues/2'>positively charged amino acids</scene>. The DNA-binding domain lies within 115 amino acid residues from this terminus. The COOH terminus includes many negatively charged, or <scene name='56/566528/Acidic_residues/5'>acidic amino acids</scene> <ref>PMID:2087220</ref>. | ||
| - | </StructureSection> | ||
==Binding Interactions in the Active Site== | ==Binding Interactions in the Active Site== | ||
Revision as of 13:09, 20 November 2013
Sandbox Single Stranded DNA-Binding Protein (SSB)
Contents |
Overview
Single-stranded DNA-binding protein (SSB) binds to single-stranded regions of DNA. This binding serves a variety of functions - it prevents the strands from hardening too early during replication, it protects the single-stranded DNA from being broken down by nucleases during repair, and it removes the secondary structure of the strands so that other enzymes are able to access them and act effectively upon the strands[1].
Single-stranded DNA (ssDNA) is utilized primarily during the course of major aspects of DNA metabolism such as replication, recombination and repair [2]. In addition to stabilizing ssDNA, SSB proteins also bind to and control the function of many other proteins that are involved in all three of these major DNA metabolic processes. During DNA replication, SSB molecules bind to the newly separated individual DNA strands, keeping the strands separated by holding them in place so that each strand can serve as a template for new DNA synthesis[3].
Structure of E. coli SSB
| |||||||||||
See Also
References
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Berg JM, Tymoczko JL, Stryer L. Biochemistry. 6th edition. New York: W H Freeman; 2006.
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002 May 14;41(19):6032-44. PMID:11993998
- ↑ Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002 May 14;41(19):6032-44. PMID:11993998
- ↑ Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002 May 14;41(19):6032-44. PMID:11993998
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Agamova KA, Gladunova ZD, Savinkin IuN. [Cytologic method in the diagnosis of precancerous conditions and early cancer of the stomach]. Lab Delo. 1988;(3):43-5. PMID:2453719
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002
- ↑ Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J Mol Biol. 2009 May 8;388(3):508-19. Epub 2009 Mar 13. PMID:19285993 doi:10.1016/j.jmb.2009.03.022
- ↑ Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Shamoo, Yousif. “Single Stranded DNA binding proteins.” ‘’Encyclopedia of Life Sciences.’’ MacMillan Publishers Ltd, Nature Publishing Group; 2002
Proteopedia Page Contributors and Editors (what is this?)
Refayat Ahsen, Rachel Craig, Michal Harel, Alexander Berchansky