Glutamate receptor (GluA2)
From Proteopedia
(→See Also) |
|||
| Line 92: | Line 92: | ||
* [[1bl8]] and [[1jq1]] and [[1jq2]] – ''Streptomyces lividans'' KcsA potassium channel<ref name="pot3" /><ref>PMID:11573095</ref>: The M1, M2 and M3 segments of GluA2's ion channel overlap remarkably well with the structurally equivalent portions KcsA. | * [[1bl8]] and [[1jq1]] and [[1jq2]] – ''Streptomyces lividans'' KcsA potassium channel<ref name="pot3" /><ref>PMID:11573095</ref>: The M1, M2 and M3 segments of GluA2's ion channel overlap remarkably well with the structurally equivalent portions KcsA. | ||
* [[Molecular Playground/Glutamate Receptor]] | * [[Molecular Playground/Glutamate Receptor]] | ||
| + | *[[Ligand Binding N-Terminal of Metabotropic Glutamate Receptors]] | ||
==References== | ==References== | ||
Revision as of 19:02, 21 November 2013
| |||||||||||
Details of Structure Featured
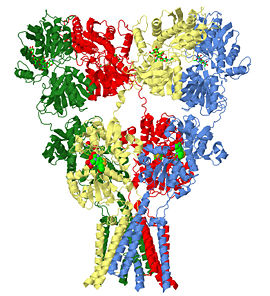
3kg2 is a 4 chains structure of sequences from Rattus norvegicus. Full crystallographic information is available from OCA. Although it is billed as the first structure of a full-length glutamate receptor, the carboxy-terminal domain is not present in the structure.
Reference for the structure
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
3D structures of glutamate receptors
Ionotropic Glutamate Receptors
See Also
- 1iiw and 1iit and 1ii5 – Prokaryotic glutamte receptor (Glur0) Apo structure and with various ligands bound, including glutmate [13]. This helped cement the notion the glutamate and potassium receptors share structural similarity and possibly evolutionary ancestry [10][11].
- 1bl8 and 1jq1 and 1jq2 – Streptomyces lividans KcsA potassium channel[12][14]: The M1, M2 and M3 segments of GluA2's ion channel overlap remarkably well with the structurally equivalent portions KcsA.
- Molecular Playground/Glutamate Receptor
- Ligand Binding N-Terminal of Metabotropic Glutamate Receptors
References
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
- ↑ Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009 Jun;16(6):631-8. Epub 2009 May 24. PMID:19465914 doi:10.1038/nsmb.1613
- ↑ Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009 Dec 16;28(24):3910-20. Epub . PMID:19910922 doi:10.1038/emboj.2009.338
- ↑ Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998 Oct 29;395(6705):913-7. PMID:9804426 doi:10.1038/27692
- ↑ 5.0 5.1 Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ Wollmuth LP, Traynelis SF. Neuroscience: Excitatory view of a receptor. Nature. 2009 Dec 10;462(7274):729-31. PMID:20010675 doi:10.1038/462729a
- ↑ Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995 Apr;18(4):161-8. PMID:7539962
- ↑ Turski L, Huth A, Sheardown M, McDonald F, Neuhaus R, Schneider HH, Dirnagl U, Wiegand F, Jacobsen P, Ottow E. ZK200775: a phosphonate quinoxalinedione AMPA antagonist for neuroprotection in stroke and trauma. Proc Natl Acad Sci U S A. 1998 Sep 1;95(18):10960-5. PMID:9724812
- ↑ Walters MR, Kaste M, Lees KR, Diener HC, Hommel M, De Keyser J, Steiner H, Versavel M. The AMPA antagonist ZK 200775 in patients with acute ischaemic stroke: a double-blind, multicentre, placebo-controlled safety and tolerability study. Cerebrovasc Dis. 2005;20(5):304-9. Epub 2005 Aug 30. PMID:16131799 doi:10.1159/000087929
- ↑ 10.0 10.1 Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995 Apr;18(4):161-8. PMID:7539962
- ↑ 11.0 11.1 Wood MW, VanDongen HM, VanDongen AM. Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proc Natl Acad Sci U S A. 1995 May 23;92(11):4882-6. PMID:7761417
- ↑ 12.0 12.1 Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998 Apr 3;280(5360):69-77. PMID:9525859
- ↑ Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999 Dec 16;402(6763):817-21. PMID:10617203 doi:10.1038/45568
- ↑ Liu YS, Sompornpisut P, Perozo E. Structure of the KcsA channel intracellular gate in the open state. Nat Struct Biol. 2001 Oct;8(10):883-7. PMID:11573095 doi:10.1038/nsb1001-883
Additional Literature and Resources
- For additional information, see: Alzheimer's Disease
- For additional information, see: Membrane Channels & Pumps
- Glutamate Receptor on the cover of Nature
- Glutamate receptor Wikipedia entry
- Glutamate Receptors page at the MRC Centre for Synaptic Plasticity at the University of Bristol
Page started with original page seeded by OCA on Wed Dec 16 11:24:54 2009 for 3kg2.
Proteopedia Page Contributors and Editors (what is this?)
Wayne Decatur, Alexander Berchansky, Michal Harel, David Canner, Nikki Hunter
Categories: Rattus norvegicus | Gouaux, E. | Rosconi, M P. | Sobolevsky, A I. | Alternative splicing | Cell membrane | Glycoprotein | Ion channel | Ion transport | Membrane | Membrane protein | Postsynaptic cell membrane | Receptor | Rna editing | Synapse | Tetramer | Transmembrane | Transport | Neuron | Neurotransmitter | Potassium Channels | RCSB PDB Molecule of the Month | Streptomyces lividans | Cabral, J M. | Chait, B T. | Cohen, S L. | Doyle, D A. | Gulbis, J M. | Kuo, A. | Mackinnon, R. | Pfuetzner, R A. | Integral membrane protein | Potassium channel | Topic Page