Sandbox Reserved 779
From Proteopedia
| Line 69: | Line 69: | ||
==References== | ==References== | ||
at least 5 | at least 5 | ||
| + | |||
| + | |||
| + | |||
| + | <big>'''example on how to put content in proteopedia:'''</big> | ||
Revision as of 13:19, 25 November 2013
[[Image:Example.jpg
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
β-Lactoglobulin
Contents |
Introduction
Function:Primary component of whey, it binds retinol and is probably involved in the transport of that molecule.
relevant background:
class of protein :Belongs to the calycin superfamily. Lipocalin family. overall function of Lipocalin family: The lipocalins are a family of proteins which transport small hydrophobic molecules such as steroids, bilins, retinoids, and lipids. Lipocalins have been associated with many biological processes, among them immune response, pheromone transport, biological prostaglandin synthesis, retinoid binding, and cancer cell interactions.
short description of protein fold: They share limited regions of sequence homology and a common tertiary structure architecture.[2][3][4][5][6] This is an eight stranded antiparallel beta-barrel with a repeated + 1 topology enclosing an internal ligand binding site.[5][4].[1] Therefore To know more abouts and the related deseases you can follow the link that leads you to the Portal to Swiss-Prot diseases and variants organisms:These proteins are found in gram negative bacteria, vertebrate cells, and invertebrate cells, and in plants.
other information: Subunit structure : Under physiological conditions beta-lactoglobulin exists as an equilibrium mixture of monomeric and dimeric forms. Subcellular location: Secreted. Tissue specificity: Synthesized in mammary gland and secreted in milk. Post-translational modification : Alternate disulfide bonds occur in equal amounts in all variants examined. Allergenic properties:Causes an allergic reaction in human. Is one of the causes of cow's milk allergy. Miscellaneous The B variant sequence is shown.
Lipocalin Proteins
The merlin-1 protein belongs to the band 4.1 superfamily of membrane-cytoskeletal linkers [2]. Within this superfamily merlin-1 is closer to ezrin,radixin and moesin (the ERM proteins). ERM proteins link adehrens junctions to the actin cytoskeleton,and are able to remodel adherens junctions during epithelial morphogenesis. They also maintain the organization of apical surfaces on the plasma membrane [3].
Structure
overall description of the structure of the protein: a. oligomeric state b. description of secondary structure c. description of active residues of the protein and where they are on the protein d. description of any ligands in the structure e. methods used to solve the structure : X-ray crystallography, NMR, EM
upload the structure (number code:2Q2M)
secondary structure elements
protein fold and how thats important for the function ligands if theres ligands the active site if relevant features of protein that are important for function zoom in on the active site, label the important active site residues, and hughlight those residues in a different color (make it look pretty)
Mechanism of action
how the protein function
include chemical structure of any relevant ligands, inhibitors, or important states in the reaction pathway.
Implications or possible application
describe any uses or application that have been made of the protein
References
at least 5
example on how to put content in proteopedia:
Structural organization
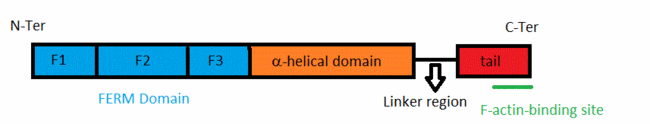
All these proteins have an about 300-residue globular plasma membrane-associated FERM domain(four-point-one ezrin, radixin, moesin).This FERM domain is a highly conserved domain and is divided into three subdomains (F1, F2, and F3). ERM proteins are composed of a FERM domain followed by a long region with a high α-helical propensity and terminating in a C-terminal domain[4].
Regulation of the activity
The acitivity of ERM proteins is caused by the association of different regions within the protein. The C-terminal tail domain contains an F-actin binding site in the last 30 residues. This domain also interacts with the FERM domain. The FERM-tail complex represents an inactive form of the protein in which membrane protein and active binding sites are masked.[5] The ERM proteins are regulated by changing from a close to an open conformation. This is due to severing of intramolecular head–tail interactions,and also of interactions between their FERM domain and α-helical domains[6].Conformational changes activate the proteins because they modify the intramolecular contacts, allowing them to bind to their partners. The FERM domain has a fundamental role because it allows ERM proteins to interact with integral proteins of the plasma membrane[7].
Regulators of the activity
Phosphorylation of a C-terminal threonine by Rho kinase as well as binding to phosphatidylinositol 4,5-bisphosphate (PIP2) and protein partners, is necessary for full activation of ERM proteins [8]. They disrupt the head to tail interactions. The phosphorylations and binding(s) determine the cellular localization and the cellular function of each specific ERM protein[9].
Merlin shares certain properties with the ERM family : they both have a subcellular localization to cortical actin structures and they both bind to adhesion receptors.These receptors are CD44 [10] and E-cadherin [11].
However Merlin-1 has some properties not shared with ERM proteins.
Specificity of merlin FERM domain
| |||||||
| 3u8z, resolution 2.64Å () | |||||||
|---|---|---|---|---|---|---|---|
| Gene: | NF2, SCH (Homo sapiens) | ||||||
| |||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
As showed in the default scene, the structure 3U8Z has in total 4 chains. These are represented by 1 sequence-unique entity. The chains A,B and C possess 9 Alpha Helices and 15 Beta Strands and the chain D has only 9 Alpha Helicesand 14 Beta Strands . You can visualize their .
Structural differences
The overall architecture of merlin is similar to that of ERM proteins. Indeed they have almost the same organization : a FERM domain,a central α-helical rod, but lack a C-terminal actin-binding site[6]. The closed complex of Merlin proteins corresponds to the tumor suppressor-active form. As the N-terminus FERM domain and C-terminus are maintained associated, Merlin is in a closed conformation and is able to promote nuclear translocation and inhibt growth[12]. More precisly,binding of the tail provokes dimerization and unfurling of the F2 motif of the FERM domain.The “closed” complex of merlin-1 is in fact an “open” dimer [4]. For more details about the probable quaternary states, see the PDBe page about the structure of 3u8z.
Merlin regulation
and Ser-518 (not in the FERM domain) phosphorylations by protein kinase A (PKA) and/or p21-activated kinase(PAK) trigger the "closed" complex [13].
Phosphorylation by PAK and PKA at Ser 518 renders the protein inactive,reducing the inhibition of cell growth.
Merlin possess a serine 10 that can also be phosphorylated by Akt. This phosphorylation directs merlin for proteasome-mediated degradation.[14].
Tumor suppressive function
The phosphoinositide 3-kinase PI3K/Akt signaling pathway is often involved in tumor proliferation. Indeed overexpression of Akt is associated with tumor development[15]. Merlin plays a fundamental role in controlling the PI3K/Akt pathway by inhibiting Akt signaling [16].
Even if the precise mechanism is not known, CD44 is absolutely required for the growth suppressive function of merlin. The protein interacts with CD44 (a transmembrane protein) but not through the same domain as ERM proteins. This interaction mediates merlin function and is regulated by the concentration of merlin in the cell and also through the concentration of hyaluronate (CD44 ligand). [17].
Applications
Nowadays, late stage melanoma is resistant to any treatment. To achieve better therapies for patients, we need to get deeper in the understanding of the signaling pathways of melanoma progression. Merlin is a target that is seriously considered because its levels and activity can be modulated through post-translational modifications[18].Phosphorylation at Ser518 of merlin inactivates its growth inhibitive activity. As we explained this phosphorylation can be achieved by cyclic AMP-dependent protein PKA and PAK1.
A mechanism that lowers merlin expression in breast cancer[19] is the phosphorylation at and . They target the protein for ubiquitination,degradation [20].
However there exist many proteins that regulate merlin expression. They may all be an useful therapetic target.Therefore further inverstigation are required to determine the pathways that involve the merlin protein.
External Resources
- See: Oncogenes & Tumor Suppressor Genes for Additional examples of oncogenes and tumor suppressor genes.
- See: Cancer for Additional Proteins involved in the disease.
- Hennigan RF, Moon CA, Parysek LM, Monk KR, Morfini G, Berth S, Brady S, Ratner N. The NF2 tumor suppressor regulates microtubule-based vesicle trafficking via a novel Rac, MLK and p38(SAPK) pathway. Oncogene. 2012 Apr 23. doi: 10.1038/onc.2012.135. PMID:22525268 doi:10.1038/onc.2012.135
- Sun CX, Robb VA, Gutmann DH. Protein 4.1 tumor suppressors: getting a FERM grip on growth regulation. J Cell Sci. 2002 Nov 1;115(Pt 21):3991-4000. PMID:12356905
- Johnson KC, Kissil JL, Fry JL, Jacks T. Cellular transformation by a FERM domain mutant of the Nf2 tumor suppressor gene. Oncogene. 2002 Sep 5;21(39):5990-7. PMID:12203111 doi:10.1038/sj.onc.1205693
- Li Q, Nance MR, Kulikauskas R, Nyberg K, Fehon R, Karplus PA, Bretscher A, Tesmer JJ. Self-masking in an intact ERM-merlin protein: an active role for the central alpha-helical domain. J Mol Biol. 2007 Feb 2;365(5):1446-59. Epub 2006 Oct 26. PMID:17134719 doi:10.1016/j.jmb.2006.10.075
- Surace EI, Haipek CA, Gutmann DH. Effect of merlin phosphorylation on neurofibromatosis 2 (NF2) gene function. Oncogene. 2004 Jan 15;23(2):580-7. PMID:14724586 doi:10.1038/sj.onc.1207142
- Mani T, Hennigan RF, Foster LA, Conrady DG, Herr AB, Ip W. FERM domain phosphoinositide binding targets merlin to the membrane and is essential for its growth-suppressive function. Mol Cell Biol. 2011 May;31(10):1983-96. doi: 10.1128/MCB.00609-10. Epub 2011 Mar , 14. PMID:21402777 doi:10.1128/MCB.00609-10
- Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000 Apr 28;101(3):259-70. PMID:10847681
References
- ↑ Martuza RL, Eldridge R. Neurofibromatosis 2 (bilateral acoustic neurofibromatosis). N Engl J Med. 1988 Mar 17;318(11):684-8. PMID:3125435 doi:http://dx.doi.org/10.1056/NEJM198803173181106
- ↑ Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al.. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993 Nov 19;75(4):826. PMID:8242753
- ↑ Brault E, Gautreau A, Lamarine M, Callebaut I, Thomas G, Goutebroze L. Normal membrane localization and actin association of the NF2 tumor suppressor protein are dependent on folding of its N-terminal domain. J Cell Sci. 2001 May;114(Pt 10):1901-12. PMID:11329377
- ↑ 4.0 4.1 4.2 Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010 Apr;11(4):276-87. doi: 10.1038/nrm2866. PMID:20308985 doi:10.1038/nrm2866
- ↑ doi: https://dx.doi.org/10.1074/jbc.274.1.170
- ↑ 6.0 6.1 Yogesha SD, Sharff AJ, Giovannini M, Bricogne G, Izard T. Unfurling of the band 4.1, ezrin, radixin, moesin (FERM) domain of the merlin tumor suppressor. Protein Sci. 2011 Oct 19. doi: 10.1002/pro.751. PMID:22012890 doi:10.1002/pro.751
- ↑ Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002 Aug;3(8):586-99. PMID:12154370 doi:10.1038/nrm882
- ↑ Fievet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, Arpin M. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J Cell Biol. 2004 Mar 1;164(5):653-9. PMID:14993232 doi:10.1083/jcb.200307032
- ↑ Mani T, Hennigan RF, Foster LA, Conrady DG, Herr AB, Ip W. FERM domain phosphoinositide binding targets merlin to the membrane and is essential for its growth-suppressive function. Mol Cell Biol. 2011 May;31(10):1983-96. doi: 10.1128/MCB.00609-10. Epub 2011 Mar , 14. PMID:21402777 doi:10.1128/MCB.00609-10
- ↑ Vaheri A, Carpen O, Heiska L, Helander TS, Jaaskelainen J, Majander-Nordenswan P, Sainio M, Timonen T, Turunen O. The ezrin protein family: membrane-cytoskeleton interactions and disease associations. Curr Opin Cell Biol. 1997 Oct;9(5):659-66. PMID:9330869
- ↑ Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003 May 1;17(9):1090-100. Epub 2003 Apr 14. PMID:12695331 doi:10.1101/gad.1054603
- ↑ Li W, Cooper J, Karajannis MA, Giancotti FG. Merlin: a tumour suppressor with functions at the cell cortex and in the nucleus. EMBO Rep. 2012 Mar;13(3):204-15. PMID:22482125
- ↑ Laulajainen M, Muranen T, Carpen O, Gronholm M. Protein kinase A-mediated phosphorylation of the NF2 tumor suppressor protein merlin at serine 10 affects the actin cytoskeleton. Oncogene. 2008 May 22;27(23):3233-43. Epub 2007 Dec 10. PMID:18071304 doi:10.1038/sj.onc.1210988
- ↑ Laulajainen M, Muranen T, Nyman TA, Carpen O, Gronholm M. Multistep phosphorylation by oncogenic kinases enhances the degradation of the NF2 tumor suppressor merlin. Neoplasia. 2011 Jul;13(7):643-52. PMID:21750658
- ↑ Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002 Jul;2(7):489-501. PMID:12094235 doi:10.1038/nrc839
- ↑ Rong R, Tang X, Gutmann DH, Ye K. Neurofibromatosis 2 (NF2) tumor suppressor merlin inhibits phosphatidylinositol 3-kinase through binding to PIKE-L. Proc Natl Acad Sci U S A. 2004 Dec 28;101(52):18200-5. Epub 2004 Dec 14. PMID:15598747 doi:0405971102
- ↑ Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, Gutmann DH, Ponta H, Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001 Apr 15;15(8):968-80. PMID:11316791 doi:10.1101/gad.189601
- ↑ Murray LB, Lau YK, Yu Q. Merlin is a negative regulator of human melanoma growth. PLoS One. 2012;7(8):e43295. doi: 10.1371/journal.pone.0043295. Epub 2012 Aug 17. PMID:22912849 doi:10.1371/journal.pone.0043295
- ↑ Morrow KA, Das S, Metge BJ, Ye K, Mulekar MS, Tucker JA, Samant RS, Shevde LA. Loss of tumor suppressor Merlin in advanced breast cancer is due to post-translational regulation. J Biol Chem. 2011 Nov 18;286(46):40376-85. doi: 10.1074/jbc.M111.250035. Epub, 2011 Sep 30. PMID:21965655 doi:10.1074/jbc.M111.250035
- ↑ Tang X, Jang SW, Wang X, Liu Z, Bahr SM, Sun SY, Brat D, Gutmann DH, Ye K. Akt phosphorylation regulates the tumour-suppressor merlin through ubiquitination and degradation. Nat Cell Biol. 2007 Oct;9(10):1199-207. Epub 2007 Sep 23. PMID:17891137 doi:10.1038/ncb1641