Sandbox Reserved 772
From Proteopedia
| Line 12: | Line 12: | ||
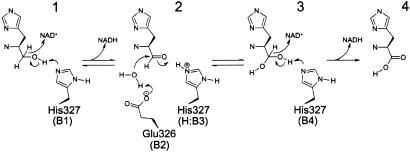
Histidinol dehydrogenase (HDH) is coded by the structural gene hisD. Histidinol dehydrogenase catalyzes the last step in the histidine biosynthetic pathway. This pathway was found in bacteria, archaebacteria, fungi, and plants. The pathway involves the conversion of L-histidinol to L-histidine with a L-histidinaldehyde intermediate <ref name="rasmol">citation</ref> | Histidinol dehydrogenase (HDH) is coded by the structural gene hisD. Histidinol dehydrogenase catalyzes the last step in the histidine biosynthetic pathway. This pathway was found in bacteria, archaebacteria, fungi, and plants. The pathway involves the conversion of L-histidinol to L-histidine with a L-histidinaldehyde intermediate <ref name="rasmol">citation</ref> | ||
| + | |||
__TOC__ | __TOC__ | ||
| Line 17: | Line 18: | ||
==General Information== | ==General Information== | ||
| - | '''Gene Name''': hisD | + | '''Gene Name''': hisD <ref name="info">citation5</ref> |
| - | '''Organism''': Escherichia coli (strain K12) | + | '''Organism''': Escherichia coli (strain K12) <ref name="info">citation5</ref> |
| - | ''' | + | '''Classification''': Oxidoreductase |
| - | '''Length''': 434 Amino Acids | + | '''Length''': 434 Amino Acids <ref name="info">citation5</ref> |
| - | '''Chains''': A, B | + | '''Chains''': A, B <ref name="cite2">cite2</ref> |
'''Molecular Weight''': | '''Molecular Weight''': | ||
| Line 39: | Line 40: | ||
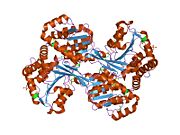
HisD is a monomer, but it functions as a homodimer. The presence of Zn2+ cation is required per monomer. Each hisD monomer is made of four domains,two larger domains (globule) and two smaller domains (extending tail), whereas the intertwined dimer possibly results from domain swapping. Two domains display a very similar incomplete Rossmann fold that suggests an ancient event of gene duplication. Residues from both monomers form the active site. The active site, residue His-327, participates in acid-base catalysis <ref name="rasmol">CITATION</ref> | HisD is a monomer, but it functions as a homodimer. The presence of Zn2+ cation is required per monomer. Each hisD monomer is made of four domains,two larger domains (globule) and two smaller domains (extending tail), whereas the intertwined dimer possibly results from domain swapping. Two domains display a very similar incomplete Rossmann fold that suggests an ancient event of gene duplication. Residues from both monomers form the active site. The active site, residue His-327, participates in acid-base catalysis <ref name="rasmol">CITATION</ref> | ||
| + | 48% helical (20 helices; 211 residues) | ||
| + | 16% beta sheet (15 strands; 73 residues) | ||
| - | '''Related Structures''': [http://proteopedia.org/wiki/index.php/1kae 1KAE] and [http://proteopedia.org/wiki/index.php/1kar 1KAR] | ||
| + | |||
| + | '''Related Structures''': [http://proteopedia.org/wiki/index.php/1kae 1KAE] and [http://proteopedia.org/wiki/index.php/1kar 1KAR] | ||
==Enzymatic Mechanism== | ==Enzymatic Mechanism== | ||
Revision as of 01:08, 3 December 2013
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
|
Histidinol Dehydrogenase
Histidinol dehydrogenase (HDH) is coded by the structural gene hisD. Histidinol dehydrogenase catalyzes the last step in the histidine biosynthetic pathway. This pathway was found in bacteria, archaebacteria, fungi, and plants. The pathway involves the conversion of L-histidinol to L-histidine with a L-histidinaldehyde intermediate [1]
Contents |
General Information
Gene Name: hisD [2]
Organism: Escherichia coli (strain K12) [2]
Classification: Oxidoreductase
Length: 434 Amino Acids [2]
Chains: A, B [3]
Molecular Weight:
Isoelectric Point:
Km:
Vmax:
Structure
HisD is a monomer, but it functions as a homodimer. The presence of Zn2+ cation is required per monomer. Each hisD monomer is made of four domains,two larger domains (globule) and two smaller domains (extending tail), whereas the intertwined dimer possibly results from domain swapping. Two domains display a very similar incomplete Rossmann fold that suggests an ancient event of gene duplication. Residues from both monomers form the active site. The active site, residue His-327, participates in acid-base catalysis [1]
48% helical (20 helices; 211 residues) 16% beta sheet (15 strands; 73 residues)
Related Structures: 1KAE and 1KAR
Enzymatic Mechanism
This bifunctional enzyme converts L-histidinol to L-histidine through a L-histidinaldehyde intermediate. [1]