Sandbox Reserved 763
From Proteopedia
| Line 101: | Line 101: | ||
Adenosine-5'-sisphosphate (ADP) | Adenosine-5'-sisphosphate (ADP) | ||
| - | Chloride ion (CL) | + | Chloride ion (<scene name='56/564039/Clligand/1'>CL</scene>) |
(3R,4S,5R)-3,4,5-trihydroxycyclohex-1-ene-1-caroxylic acid (SKM) | (3R,4S,5R)-3,4,5-trihydroxycyclohex-1-ene-1-caroxylic acid (SKM) | ||
Revision as of 01:44, 4 December 2013
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
Contents |
Shikimate Kinase
|
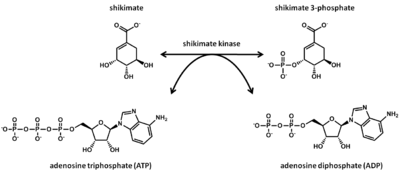
Shikimate kinase is an enzyme which participates in the fifth step of the shikimate pathway. The functional role shikimate kinase plays in this pathway is to catalyze the ATP-dependent phosphorylation of shikimate into shikimate 3-phosphate (3-phosphoshikimate). This aids in the synthesis of chorismate, which is the precursor to aromatic amino acids and secondary metabolites. Shikimate kinase is a protein kinase, an enzyme which phosphorylates a protein leading to a functional change in the phosphorylated protein, of the transferase class. A transferase acts to transfer a functional group from the donor to acceptor molecule. The protein fold consists of 9 helices and 5 strands. The length of shikimate kinase ranges from 161-214 residues and it is found in the cytoplasm. This is a subclass of alpha/beta proteins, meaning an alpha/beta domain exists. The subunit is that of a monomer. This enzyme consists of the CORE, LED, and substrate-binding domains. ATP is the cosubstrate while magnesium ion is the cofactor. Though the shikimate pathway is present in microorganisms and plants, shikimate kinase is found only in the bacteria taxon. This enzyme is found predominantly in the organism Mycobacterium tuberculosis, but also in Helicobacter pylori, Bacteroides thetaiotaomicron, Campylobacter jejuni, Aquifex aeolicus, Coxiella burnetii, Arabidopsis thaliana. This enzyme is a protein target for rational drug design against tuberculosis, which holds great potential due to shikimate kinase being absent in mammals.
Structure
The crystal structure of 2iyq from the Protein Data Bank [1] showing shikimate kinase from Mycobacterium tuberculosis complexed with ADP and shikimate is shown to the right as the . In addition, the 2D model of 2gij, showing the space filling details of the asymmetric unit of MtSK, is shown to the left.
3D Structures in Different Organisms
Mycobacterium tuberculosis
2g1j - MtSK
2iyt - MtSK (unliganded, open lid)
1iyw - MtSK (open lid) + ATP
2g1k, 2iyr, 2iyx - MtSK + shikimate
2iys - MtSK (open lid) + shikimate
2iyy - MtSK + shikimate-3-phosphate
2iys - MtSK (open lid) + shikimate
2iyu, 2iyv - MtSK (open lid) + ADP
1l4u, 1l4y, 1u8a, 2dft - MtSK + ADP
2iyw - MtSK (open lid) + ATP
3baf - MtSK + AMP-PNP
2dfn, 1we2, 2iyq, 1u8a - MtSK + ADP + shikimate
2iyz - MtSK + ADP + shikimate-3-phosphate
4bqs - MtSK + ADP + shikimic acid derivative
1zyu - MtSK + AMPPCP + shikimate
Helicobacter pylori
1zuh - HpSK
3mrs - HpSK (mutant)
1zui - HpSK + shikimate
3n2e - HpSK (mutant) + inhibitor
3muf - HpSK + ADP + shikimate-3-phosphate
Other Organisms
2pt5 - SK - Aquifex Aeolicus
3trf - SK - Coxiella burnetii
3vaa - SK - Bacteroides thetaiotaomicron
1via - SK - Campylobacter jejuni
3nwj - SK - Arabidopsis thaliana
Secondary Structural Elements
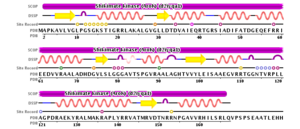
Using 2iyq as an example, the secondary structure of shikimate kinase can be observed in terms of strands and helices shown . There are 5 strands, shown in yellow, and 9 helices, shown in pink. Shown to the left is a 2D depiction of the secondary structural content of shikimate kinase (strands are yellow, helices are pink) and corresponding residues.
Oligomeric State
Through experiments involving size exclusion liquid chromatography and gel filtration chromatography, the oligomeric state of homogeneous MtSK was found. The molecular mass was found to be 20.7 kDa and when compared to the suggested value of 18.5 kDa, it was found that MtSK is a monomer in solution.[2]
Active Residues
Using 2iyq as an example, the active residues were binding in shikimate kinase occurs is shown to the left. There are three domains present in Shikimate kinase: the CORE domain, and substrate-binding domain, and LID domain. Each domain plays a role in binding and is related to the active residues. The binding site for nucleotides is in the CORE, shikimate binds in the substrate-binding domain, and once ATP or shikimate binds, the LID domain closes over the active site.[3]
Ligands
Ligands associated with shikimate kinase include ADP, CL, SKM, and TRS.
Adenosine-5'-sisphosphate (ADP)
Chloride ion ()
(3R,4S,5R)-3,4,5-trihydroxycyclohex-1-ene-1-caroxylic acid (SKM)
2-amino-2-hydroxymethyl-propane-1,3-diol (TRS)
Protein Fold
Protein folding from a random coil to native state with correct 3D structure is essential for proper protein function. The primary sequence of amino acids determines the folded structure.
Shikimate kinase is a convenient protein to use in protein folding studies. This is because it is one of the smallest kinases and is a monomeric enzyme lacking disulfide bonds. It has been shown that the proposed refolding model includes a rapid hydrophobic collapse and then a slower secondary structure formation.[4]
Look into how the protein fold could be disrupted (ie pH, temperature studies etc)
Methods Used to Solve
Mechanism of Action
Describe how protein functions. Structures of Ligands/inhibitors/important steps in reaction pathway.
The substrates involved with shikimate kinase are shikimate and ATP. The products involved are shikimate 3-phosphate and ADP.
Implications or Possible Application
Medical importance - related disease, if used as drug target.
If protein is found in other organisms.
Other uses (ie antibiotics etc)
References
- ↑ 1.0 1.1 1.2 Protein Data Bank http://www.rcsb.org/pdb/explore.do?structureId=2IYQ
- ↑ The Mode of Action of Recombinant Mycobacterium tuberculosis Shikimate Kinase: Kinetics and Thermodynamics Analyses http://www.plosone.org/article/info:doi/10.1371/journal.pone.0061918
- ↑ UniProt http://www.uniprot.org/uniprot/P0A4Z2
- ↑ The refolding of type II shikimate kinase from Erwinia chrysanthemi after denaturation in urea. European Journal of Biochemistry http://onlinelibrary.wiley.com/doi/10.1046/j.1432-1033.2002.ejb.02862.x/pdf