Sandbox Reserved 764
From Proteopedia
(→EPSP Synthase Structure) |
(→EPSP Synthase Structure) |
||
| Line 34: | Line 34: | ||
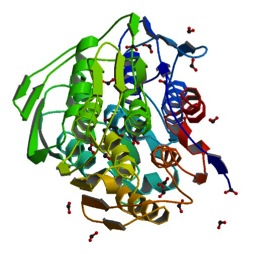
<Structure load='2AA9' size='500' frame='true' align='right' caption='EPSP synthase liganded with shikimate-3-phosphate' scene='56/564040/Lys22/1' /> | <Structure load='2AA9' size='500' frame='true' align='right' caption='EPSP synthase liganded with shikimate-3-phosphate' scene='56/564040/Lys22/1' /> | ||
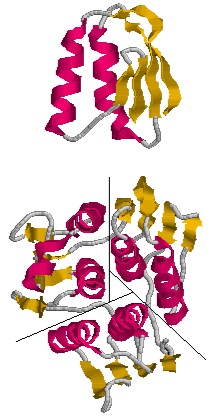
| - | The three-dimensional structure of EPSP synthase in ''Escherichia coli'' was determined using X-ray crystallography. The enzyme has two distinct domains, each of which have a radius of approximately 25 Angstroms <ref>PMID:11607190</ref>. The two | + | The three-dimensional structure of EPSP synthase in ''Escherichia coli'' was determined using X-ray crystallography. The enzyme has two distinct domains, each of which have a radius of approximately 25 Angstroms <ref>PMID:11607190</ref>. The two domains are joined together by two crossover chain segments with both the amino and carboxyl terminus of the polypeptide chain located in the C-terminal domain. The domains are formed by protein folding subunits consisting of the following <scene name='56/564040/Epsp_synthase/2'>secondary structures</scene>: two parallel helices and four beta sheets <ref>PMID:11607190</ref>. Each domain is formed from three of these subunits, forming the threefold axis of symmetry shown below in Figure 3. Three alpha helices are positioned around the axis of each domain to form its core. The domain surfaces consist of three beta-sheets and three parallel helices that are all solvent accessible <ref>PMID:11607190</ref>. The beta sheets can be found in both the parallel and anti-parallel positions. The alpha helices are positioned in a parallel fashion that establishes equivalent chain polarities throughout the enzyme. It is believed that the helix dipole establishes optimal interaction between the enzyme and charged phosphate groups of the substrates <ref> W. G. J. Hol, P. T. van Duijnen & H. J. C. The [alpha]-helix dipole and the properties of proteins: Nature 273, 443-446 (8 June 1978).[http://dx.doi.org/10.1038/273443a0 DOI:10.1038/273443a0</ref>. Studies suggest that these helical dipole efffects create a cationic well that draws the anionic substrates into the active site located in the interdomain cleft. |
<scene name='56/564040/Secondary_elements/2'>bound</scene> | <scene name='56/564040/Secondary_elements/2'>bound</scene> | ||
Revision as of 00:12, 7 December 2013
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
Contents |
5-Enolpyruvylshikimate-3-phosphate synthase
Organism: Plants and microorganisms
Subcellular location: Cytoplasm
Formula weight: approximately 47,000 Da
Classification: Transferase
Length: 427 residues
Domains: 2
Ligands: Shikimate, Formic Acid
Symbol: EPSP synthase or EPSPS
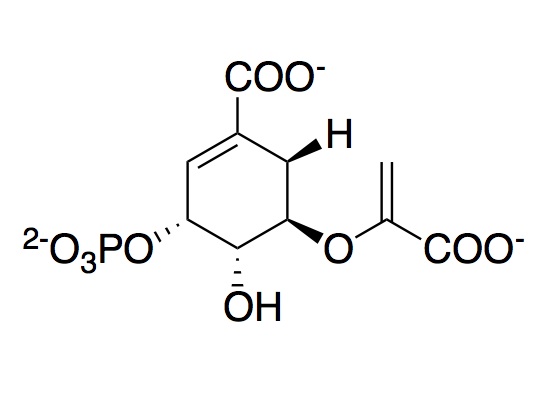
The monomeric enzyme 5-Enolpyruvlyshikimate-3-phosphate synthase (EPSP synthase) is an enzyme involved in the shikimate pathway found only in plants and microorganisms. The shikimate pathway is essential for the biosynthesis of chorismate, a molecule that is a precursor to majority of the aromatic compounds produced in the cell, including the aromatic amino acids (L-tyrosine, L-phenylalanine, L-tryptophan) [1]. These aromatic compounds are essential for the synthesis of proteins thereby making chorismate and the shikimate pathway essential for plants and microorganisms. EPSP synthase is a transferase or an enzyme that catalyzes the transfer of a specific functional group from one molecule to another. EPSP synthase catalyzes the transfer of the enolpyruvyl group from phosphonenolpyruvate (PEP) to shikimate-3-phosphate (S3P) to form 5-enolpyruvylshikimate-3-phosphate (EPSP) and inorganic phosphate. EPSP is converted into chorismate, which is then introduced into synthesis pathways of aromatic compounds [2]. EPSP synthase has two distinct globular domains, composed of beta sheets and alpha helices [3]. Each of the domains are composed of three protein folding subunits. The subunits contain two alpha helices and four beta sheets [4]. As shown in Figure 3 below, each domain has six alpha helices and twelve beta sheets. The two domains, which are linked together by two crossover chain segments, exist in an open conformation until the binding of shikimate-3-phosphate, which is the enzyme's binding substrate [5]. Upon the binding of shikimate-3-phosphate, the domains close together to form the active site in an interdomain cleft. Phosphoenolpyruvate then enters the active site enabling EPSP synthase to catalyze the transfer of the enolpyruvyl group through an addition-elimination reaction. EPSP synthase is attractive for drug research because of its potential to serve as a selective target for antimicrobial drugs [6]. Because EPSP synthase is found only in plants and microorganisms, its use in antimicrobial drugs will limit the harmful side effects for humans. antimicrobial drugs
EPSP Synthase Structure
|
The three-dimensional structure of EPSP synthase in Escherichia coli was determined using X-ray crystallography. The enzyme has two distinct domains, each of which have a radius of approximately 25 Angstroms [7]. The two domains are joined together by two crossover chain segments with both the amino and carboxyl terminus of the polypeptide chain located in the C-terminal domain. The domains are formed by protein folding subunits consisting of the following : two parallel helices and four beta sheets [8]. Each domain is formed from three of these subunits, forming the threefold axis of symmetry shown below in Figure 3. Three alpha helices are positioned around the axis of each domain to form its core. The domain surfaces consist of three beta-sheets and three parallel helices that are all solvent accessible [9]. The beta sheets can be found in both the parallel and anti-parallel positions. The alpha helices are positioned in a parallel fashion that establishes equivalent chain polarities throughout the enzyme. It is believed that the helix dipole establishes optimal interaction between the enzyme and charged phosphate groups of the substrates [10]. Studies suggest that these helical dipole efffects create a cationic well that draws the anionic substrates into the active site located in the interdomain cleft.
EPSP Synthase Mechanism
EPSP synthase catalyzes the reversible transfer of the enolpyruvyl group from phosphoenolpyruvate (PEP) to shikimate-3-phosphate (S3P) to form the products EPSP and inorganic phosphate. Fluorescence studies show that both S3P and PEP are capable of binding to free EPSP synthase [11]. However, it has been found that PEP preferentially interacts with the E-S3P complex [12]. Steady-state and pre-steady-state kinetic studies indicate that the kinetically preferred reactions pathway occurs with S3P binding to EPSP synthase first, followed by PEP [13]. Chemical modification studies on EPSPS indicate that Lys, Arg, and His residues are essential for activity of the enzyme. The reactive Lysine and Arginine residues have been identified as and . Mutation of Lys-22 to Arg does not affect the activity of the enzyme. However, mutation of Lys-22 to either Ala or Glu leads to complete loss of enzyme activity; therefore suggesting that the cationic nature of the Lysine side chain plays an important role in the activity of EPSP synthase [14]. It is speculated that both Lys-22 and Arg-27 constitute a part of the active site recognizing the anionic residues on the substrates. The following residues were proposed to be involved in the enzyme mechanism:
-E341 may as a proton donor for the C-3 of PEP. In the x-ray structure, E341 is located close to the C-2 position of PEP, and thus the carboxyl oxygen of E341 could stabilize PEP oxonium ion.
-His-385 of which is only 2.85 Å away from O of E341 may function as proton source for E341.
-Two residues, D313 and K22 may act as a proton acceptor of the 5-OH of S3P which should be deprotonated to attack the C-2 of PEP (24) and interacts only with those two residues (Scheme 1).
-R100, D242, and D384 residues are not involved in substrate binding/catalysis, but mutagenisis results show that they are required. It is proposed that they may play a role in domain closure or stabilize the closed conformation.
Implications
The implications in studying EPSP synthase involve the fact that this enzyme is not present in humans but is essential for bacteria and apicomplexan parasites. This makes EPSP synthase a selective target for antimicrobial drugs and decreases possible negative impacts of drugs in humans.
References
- ↑ Lewis J, Johnson KA, Anderson KS. The catalytic mechanism of EPSP synthase revisited. Biochemistry. 1999 Jun 1;38(22):7372-9. PMID:10353849 doi:http://dx.doi.org/10.1021/bi9830258
- ↑ Lewis J, Johnson KA, Anderson KS. The catalytic mechanism of EPSP synthase revisited. Biochemistry. 1999 Jun 1;38(22):7372-9. PMID:10353849 doi:http://dx.doi.org/10.1021/bi9830258
- ↑ Stallings WC, Abdel-Meguid SS, Lim LW, Shieh HS, Dayringer HE, Leimgruber NK, Stegeman RA, Anderson KS, Sikorski JA, Padgette SR, Kishore GM. Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5046-50. PMID:11607190
- ↑ Stallings WC, Abdel-Meguid SS, Lim LW, Shieh HS, Dayringer HE, Leimgruber NK, Stegeman RA, Anderson KS, Sikorski JA, Padgette SR, Kishore GM. Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5046-50. PMID:11607190
- ↑ Marques MR, Pereira JH, Oliveira JS, Basso LA, de Azevedo WF Jr, Santos DS, Palma MS. The inhibition of 5-enolpyruvylshikimate-3-phosphate synthase as a model for development of novel antimicrobials. Curr Drug Targets. 2007 Mar;8(3):445-57. PMID:17348837
- ↑ Marques MR, Pereira JH, Oliveira JS, Basso LA, de Azevedo WF Jr, Santos DS, Palma MS. The inhibition of 5-enolpyruvylshikimate-3-phosphate synthase as a model for development of novel antimicrobials. Curr Drug Targets. 2007 Mar;8(3):445-57. PMID:17348837
- ↑ Stallings WC, Abdel-Meguid SS, Lim LW, Shieh HS, Dayringer HE, Leimgruber NK, Stegeman RA, Anderson KS, Sikorski JA, Padgette SR, Kishore GM. Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5046-50. PMID:11607190
- ↑ Stallings WC, Abdel-Meguid SS, Lim LW, Shieh HS, Dayringer HE, Leimgruber NK, Stegeman RA, Anderson KS, Sikorski JA, Padgette SR, Kishore GM. Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5046-50. PMID:11607190
- ↑ Stallings WC, Abdel-Meguid SS, Lim LW, Shieh HS, Dayringer HE, Leimgruber NK, Stegeman RA, Anderson KS, Sikorski JA, Padgette SR, Kishore GM. Structure and topological symmetry of the glyphosate target 5-enolpyruvylshikimate-3-phosphate synthase: a distinctive protein fold. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5046-50. PMID:11607190
- ↑ W. G. J. Hol, P. T. van Duijnen & H. J. C. The [alpha]-helix dipole and the properties of proteins: Nature 273, 443-446 (8 June 1978).[http://dx.doi.org/10.1038/273443a0 DOI:10.1038/273443a0
- ↑ Kishore GM, Shah DM. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627-63. PMID:3052285 doi:http://dx.doi.org/10.1146/annurev.bi.57.070188.003211
- ↑ Kishore GM, Shah DM. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627-63. PMID:3052285 doi:http://dx.doi.org/10.1146/annurev.bi.57.070188.003211
- ↑ Lewis J, Johnson KA, Anderson KS. The catalytic mechanism of EPSP synthase revisited. Biochemistry. 1999 Jun 1;38(22):7372-9. PMID:10353849 doi:http://dx.doi.org/10.1021/bi9830258
- ↑ Kishore GM, Shah DM. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627-63. PMID:3052285 doi:http://dx.doi.org/10.1146/annurev.bi.57.070188.003211