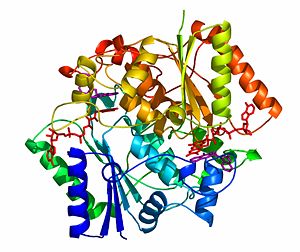
NADH quinone oxidoreductase (NQO1) with inhibitor dicoumarol
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| + | <StructureSection load='2f1o.pdb' size='450' frame='true' align='right' scene='2f1o/Com_view/2' caption='NADPH dehydrogenase complex with FAD and dicoumarol [[2f1o]]'> | ||
== The crystal structure of NADH quinone oxidoreductase (NQO1) in complex with its potent inhibitor dicoumarol == | == The crystal structure of NADH quinone oxidoreductase (NQO1) in complex with its potent inhibitor dicoumarol == | ||
| - | [[Image:NQO_Dic copy.jpg|border| | + | [[Image:NQO_Dic copy.jpg|border|left|300px]] |
| - | + | {{Clear}} | |
[http://en.wikipedia.org/wiki/NAD(P)H_dehydrogenase_(quinone) NAD(P)H quinone oxidoreductase 1] (NQO1, [http://www.expasy.org/cgi-bin/nicezyme.pl?1.6.5.2 EC 1.6.5.2]) is a ubiquitous [http://medical-dictionary.thefreedictionary.com/flavoenzyme flavoenzyme] that [http://en.wikipedia.org/wiki/Catalysis catalyzes] two electron reduction of [http://en.wikipedia.org/wiki/Quinone quinones] to [http://en.wikipedia.org/wiki/Hydroquinone hydroquinones] utilizing [[NAD(P)H]] as an [http://en.wikipedia.org/wiki/Electron_donor electron donor]. | [http://en.wikipedia.org/wiki/NAD(P)H_dehydrogenase_(quinone) NAD(P)H quinone oxidoreductase 1] (NQO1, [http://www.expasy.org/cgi-bin/nicezyme.pl?1.6.5.2 EC 1.6.5.2]) is a ubiquitous [http://medical-dictionary.thefreedictionary.com/flavoenzyme flavoenzyme] that [http://en.wikipedia.org/wiki/Catalysis catalyzes] two electron reduction of [http://en.wikipedia.org/wiki/Quinone quinones] to [http://en.wikipedia.org/wiki/Hydroquinone hydroquinones] utilizing [[NAD(P)H]] as an [http://en.wikipedia.org/wiki/Electron_donor electron donor]. | ||
NQO1 is a homo-dimer that functions via a “ping pong” mechanism. NAD(P)H binds to NQO1, reduces the [http://en.wikipedia.org/wiki/FAD FAD] co-factor and is then released, allowing the quinone substrate to bind the enzyme and to be reduced. The NAD(P)H and the quinone binding sites of NQO1 have a significant overlap, thus providing a molecular basis for this “ping pong” mechanism. | NQO1 is a homo-dimer that functions via a “ping pong” mechanism. NAD(P)H binds to NQO1, reduces the [http://en.wikipedia.org/wiki/FAD FAD] co-factor and is then released, allowing the quinone substrate to bind the enzyme and to be reduced. The NAD(P)H and the quinone binding sites of NQO1 have a significant overlap, thus providing a molecular basis for this “ping pong” mechanism. | ||
Certain [http://en.wikipedia.org/wiki/Coumarin coumarins], [http://en.wikipedia.org/wiki/Flavones flavones] and the reactive dye cibacron blue are [http://en.wikipedia.org/wiki/Competitive_inhibition competitive inhibitors] of NQO1 activity, which compete with NAD(P)H for binding to NQO1. [[Dicoumarol]] (3-3’–methylene-bis (4-hydroxycoumarin)),[[Image:Figure3 copy.jpg|border|center|300px]] is the most potent competitive inhibitor of NQO1. Dicoumarol competes with NAD(P)H for binding to NQO1 and prevents the [http://en.wikipedia.org/wiki/Electron_transfer electron transfer] to FAD. | Certain [http://en.wikipedia.org/wiki/Coumarin coumarins], [http://en.wikipedia.org/wiki/Flavones flavones] and the reactive dye cibacron blue are [http://en.wikipedia.org/wiki/Competitive_inhibition competitive inhibitors] of NQO1 activity, which compete with NAD(P)H for binding to NQO1. [[Dicoumarol]] (3-3’–methylene-bis (4-hydroxycoumarin)),[[Image:Figure3 copy.jpg|border|center|300px]] is the most potent competitive inhibitor of NQO1. Dicoumarol competes with NAD(P)H for binding to NQO1 and prevents the [http://en.wikipedia.org/wiki/Electron_transfer electron transfer] to FAD. | ||
In addition to its role in the detoxification of quinones, NQO1 is also a 20S proteasome-associated protein that plays an important role in the stability of the [http://en.wikipedia.org/wiki/Tumor_suppressor_gene tumor suppressor] p53 and several other short-lived proteins including [[p73α]] and ornithine decarboxylase (ODC, ''i.e.'' [[7odc]]). NQO1 binds and stabilizes [[p53]], protecting p53 from [http://en.wikipedia.org/wiki/Proteasome#Ubiquitin-independent_degradation ubiquitin-independent 20S proteasomal degradation]. Dicoumarol and several other inhibitors of NQO1 activity, which compete with NADH for binding to NQO1, disrupt the binding of NQO1 to p53 and induce ubiquitin-independent p53 degradation. | In addition to its role in the detoxification of quinones, NQO1 is also a 20S proteasome-associated protein that plays an important role in the stability of the [http://en.wikipedia.org/wiki/Tumor_suppressor_gene tumor suppressor] p53 and several other short-lived proteins including [[p73α]] and ornithine decarboxylase (ODC, ''i.e.'' [[7odc]]). NQO1 binds and stabilizes [[p53]], protecting p53 from [http://en.wikipedia.org/wiki/Proteasome#Ubiquitin-independent_degradation ubiquitin-independent 20S proteasomal degradation]. Dicoumarol and several other inhibitors of NQO1 activity, which compete with NADH for binding to NQO1, disrupt the binding of NQO1 to p53 and induce ubiquitin-independent p53 degradation. | ||
| - | |||
| - | {{Clear}} | ||
| - | <StructureSection load='2f1o.pdb' size='500' frame='true' align='right' scene='2f1o/Com_view/2' caption='NADPH dehydrogenase complex with FAD and dicoumarol [[2f1o]]'> | ||
The crystal structure of human NQO1 in complex with dicoumarol was determined at 2.75 Å resolution ([[2f1o]]). NQO1 is a <scene name='2f1o/Com_view/6'>physiological homodimer</scene> composed of two interlocked monomers. <scene name='2f1o/Com_view/7'>Two catalytic sites</scene> are formed and are present at the dimer interface (<font color='red'><b>FAD is colored red</b></font> and <font color='blue'><b>dicoumarol is colored blue</b></font>). Therefore, each from these two <scene name='2f1o/Active_site/3'>dicoumarol-hNQO1 binding sites</scene> is formed by both monomers. <font color='cyan'><b>Dicoumarol is colored cyan</b></font>, <font color='orange'><b>FAD in orange</b></font>, nitrogens and oxygens are shown in [http://en.wikipedia.org/wiki/CPK_coloring CPK colors]. NQO1 <font color='blueviolet'><b>chain A is colored blueviolet</b></font> and <font color='lime'><b>chain C in lime</b></font>. NQO1 residues, participating in ligand interactions, are shown as stick representation and are labeled (A and C refer to the NQO1 chains). H-bonds are shown by dashed lines with their distances. | The crystal structure of human NQO1 in complex with dicoumarol was determined at 2.75 Å resolution ([[2f1o]]). NQO1 is a <scene name='2f1o/Com_view/6'>physiological homodimer</scene> composed of two interlocked monomers. <scene name='2f1o/Com_view/7'>Two catalytic sites</scene> are formed and are present at the dimer interface (<font color='red'><b>FAD is colored red</b></font> and <font color='blue'><b>dicoumarol is colored blue</b></font>). Therefore, each from these two <scene name='2f1o/Active_site/3'>dicoumarol-hNQO1 binding sites</scene> is formed by both monomers. <font color='cyan'><b>Dicoumarol is colored cyan</b></font>, <font color='orange'><b>FAD in orange</b></font>, nitrogens and oxygens are shown in [http://en.wikipedia.org/wiki/CPK_coloring CPK colors]. NQO1 <font color='blueviolet'><b>chain A is colored blueviolet</b></font> and <font color='lime'><b>chain C in lime</b></font>. NQO1 residues, participating in ligand interactions, are shown as stick representation and are labeled (A and C refer to the NQO1 chains). H-bonds are shown by dashed lines with their distances. | ||
Revision as of 14:13, 23 December 2013
| |||||||||||
3D structures of NQ01
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- Faig M, Bianchet MA, Talalay P, Chen S, Winski S, Ross D, Amzel LM. Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release. Proc Natl Acad Sci U S A. 2000 Mar 28;97(7):3177-82. PMID:10706635 doi:http://dx.doi.org/10.1073/pnas.050585797
- Asher G, Dym O, Tsvetkov P, Adler J, Shaul Y. The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol. Biochemistry. 2006 May 23;45(20):6372-8. PMID:16700548 doi:10.1021/bi0600087
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Orly Dym, Michal Harel, Jaime Prilusky, Moshe Ben-David, Joel L. Sussman, David Canner, Eric Martz