Introduction :
Gyrase is the only prokaryote DNA topoisomerase II able to introduce negative supercoils in the DNA in order to remove positive supercoils. It catalyses the hydrolysis of two phosphodiester bonds in a DNA segment (called G segment). Then, thanks to ATP dependant conformation changes it enables the passage of another segment (the T segment) through the break, and then religates the broken segment. Gyrase acts prior to the replication (before the replication fork) or other mecanisms requiring loose DNA.
In abscence of ATP, like other topoisomerases II, gyrase only relaxes supercoils.
Gyrase is coded by two differents contiguous genes gyrA and GgyrB as it is a 350 kDa A2B2 heterotetramers of two A proteins and two B proteins.
The A protein breaks and religates DNA . The B protein has ATPase activity
Structure
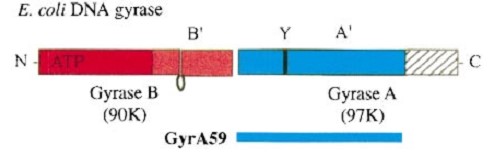
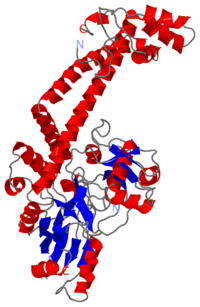
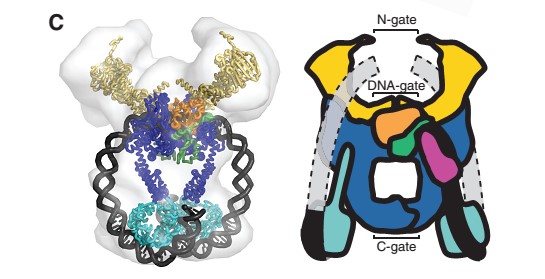
The A protein breaks and religates DNA as the DNA cleavage core and the CTD lies on it.
The gyrA 59kDa N-ter domain is called Breakage and reunion domain It is composed of two domains at the head region : a winged-helix-turn-helix domain or winged helix domain (WHD) where lies the catalytic tyrosines and a tower domain with alpha/beta structure.
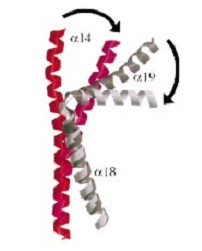
And a single domain with a helical core at the tail region. Two long helices (a14 anda18) emanate from this core and connect, together with the C-terminal helix (a19), the head and tail fragments.
Image:GyrA59.jpg
The three connecting helices (a14,a18 anda19) adopt very different conformations, leading to large quaternary movements involving a single hinge-point within the helices and rigid body movements of
the head fragments.
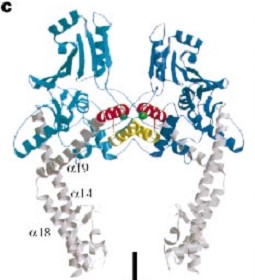
The tail is structurally conserved although large surface loops emanating from different points give it a different outward appearance It forms a heart-shaped homodimer with two protein interfaces, the DNA- and C-gates. GyrA59 is the minimal fragment of the A-subunit which, when complexed with the B-subunit, has DNA-cleavage activity.
The remaining 30–35 kDa comprising the C-terminal domain (CTD) of GyrA shows a domain forming a b-pinwheel with a positively charged amino-acid perimeter. This carboxy-terminal domain of GyrA (cyan) is required for the introduction of DNA supercoils).
The B protein has the ATPase domain and the Toprim fold on it. Two ATPase domains dimerize to form a closed conformation. The Toprim fold is a Rossmann fold (beta-alpha-beta-alpha-beta) that contains three invariant acidic residues that coordinate magnesium ions involved in DNA cleavage and DNA religation.
The central core of the protein is made out of the association of GyrA and Gyr B with the Toprim fold and the DNA-binding domain leading to a tower domain. A coiled-coil region leads to a C-terminal domain that forms the main dimer interface
The Toprim domain is flexible and that this flexibility can allow the Toprim domain to coordinate with
the WHD to form a competent cleavage complex
DNA is bent by ~150 degrees through an invariant isoleucine (in topoisomerase II it is I833 and in gyrase it is I172)
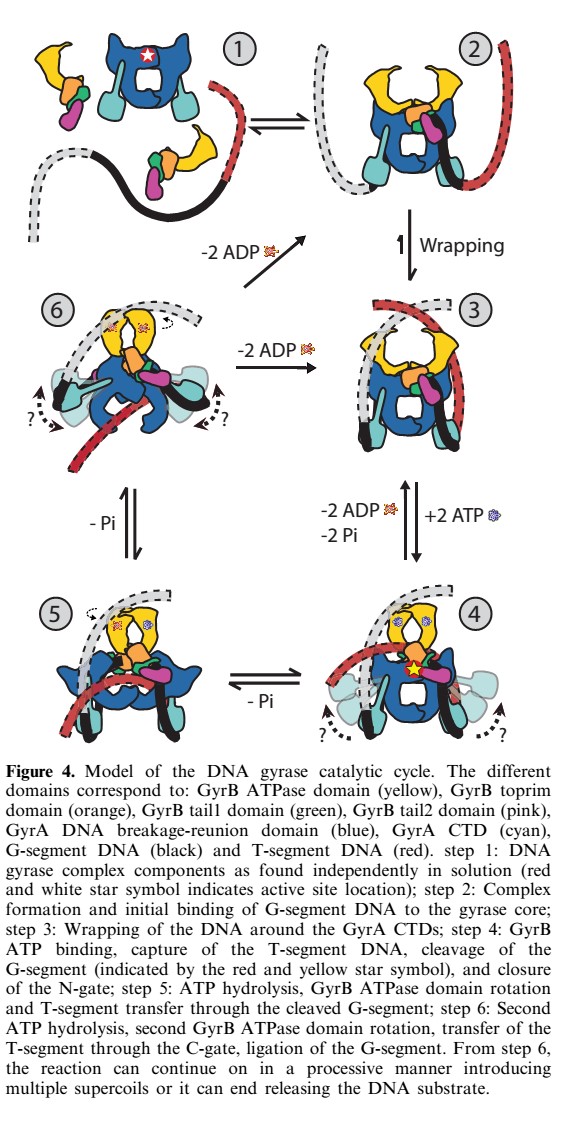
Mecanism
40-bp of duplex DNA, the G-segment, bind to the core of the enzyme and are cleaved by the active site tyrosines, while another DNA duplex, the T-segment, is captured through the ATP-induced dimerization of a protein gate, the N-gate. After passage through the transiently broken G-segment (DNA gate), the T-segment exits the protein through another protein gate, the C-gate. ATP hydrolysis and release reset the conformation of the enzyme and DNA to their initial state, poised for another strand-passage event or release of the DNA.
N- and C-gates that open or close to allow T-segment transport through both theprotein and the cleaved G-segment.
Reaction
The cleavage of DNA is achieved by a transesterification reaction between the tyrosines (Tyr122) and the target phosphoryl groups on opposing strands of the DNA backbone, resulting in the tyrosine being covalentlty attached to the 5' end of the cleaved segment with a 4-base overhang.
The active site tyrosines (Tyr 122) are on loops at either end of the dimer interface, 30Å apart, and sit at the ends of strongly basic grooves created by the dimer-related monomers.
The gyrase structure reveals a new cluster of conserved residues, juxtaposing Tyr 122 and Arg 121 from one monomer and His 80, Arg 32 and Lys 42 from the other monomer. This cluster may form the active site of the breakage–reunion reaction, with the other conserved positive charges (Arg 46 and Arg 47) anchoring the non-covalently bound 3' end of the cleaved DNA.