We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 824
From Proteopedia
(Difference between revisions)
| Line 58: | Line 58: | ||
The residues responsible for this interaction are localized in <scene name='56/568022/Hsrp54m_rna_bindv3/1'>helices 4, 5, 6 and 7</scene>, but most particularly in helices 5 and 6. | The residues responsible for this interaction are localized in <scene name='56/568022/Hsrp54m_rna_bindv3/1'>helices 4, 5, 6 and 7</scene>, but most particularly in helices 5 and 6. | ||
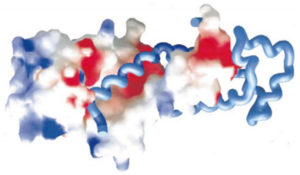
A large part of the top is therefore positively charged to bind with the negatively charged 7S RNA. | A large part of the top is therefore positively charged to bind with the negatively charged 7S RNA. | ||
| - | [[Image:Proteopedia charge.png|300px|center|thumb| | + | [[Image:Proteopedia charge.png|300px|center|thumb| In the left side we can the SRP RNA part in blue, representing the positively charged part of SRP54M. See reference 1 for more details.]] |
It was also shown that the SRP54M - SRP RNA interaction is highly dependent of the structural integrity of the <scene name='56/568022/Hsrp54m_core_and_h1/1'>SRP54M Core structure</scene>. | It was also shown that the SRP54M - SRP RNA interaction is highly dependent of the structural integrity of the <scene name='56/568022/Hsrp54m_core_and_h1/1'>SRP54M Core structure</scene>. | ||
| Line 78: | Line 78: | ||
[[1mfq]] – hSRP19 + hSRP54 M domain + 7S RNA S domain | [[1mfq]] – hSRP19 + hSRP54 M domain + 7S RNA S domain | ||
| - | == | + | ==References== |
# Clemons, W M, Jr, K Gowda, S D Black, C Zwieb, and V Ramakrishnan. “Crystal Structure of the Conserved Subdomain of Human Protein SRP54M at 2.1 A Resolution: Evidence for the Mechanism of Signal Peptide Binding.” Journal of Molecular Biology 292, no. 3 (September 24, 1999): 697–705. doi:10.1006/jmbi.1999.3090. | # Clemons, W M, Jr, K Gowda, S D Black, C Zwieb, and V Ramakrishnan. “Crystal Structure of the Conserved Subdomain of Human Protein SRP54M at 2.1 A Resolution: Evidence for the Mechanism of Signal Peptide Binding.” Journal of Molecular Biology 292, no. 3 (September 24, 1999): 697–705. doi:10.1006/jmbi.1999.3090. | ||
# Halic, Mario, Thomas Becker, Martin R. Pool, Christian M. T. Spahn, Robert A. Grassucci, Joachim Frank, and Roland Beckmann. “Structure of the Signal Recognition Particle Interacting with the Elongation-Arrested Ribosome.” Nature 427, no. 6977 (February 26, 2004): 808–814. doi:10.1038/nature02342. | # Halic, Mario, Thomas Becker, Martin R. Pool, Christian M. T. Spahn, Robert A. Grassucci, Joachim Frank, and Roland Beckmann. “Structure of the Signal Recognition Particle Interacting with the Elongation-Arrested Ribosome.” Nature 427, no. 6977 (February 26, 2004): 808–814. doi:10.1038/nature02342. | ||
# Huang, Qiaojia, Sayran Abdulrahman, Jiaming Yin, and Christian Zwieb. “Systematic Site-Directed Mutagenesis of Human Protein SRP54: Interactions with Signal Recognition Particle RNA and Modes of Signal Peptide Recognition.” Biochemistry 41, no. 38 (September 24, 2002): 11362–11371. | # Huang, Qiaojia, Sayran Abdulrahman, Jiaming Yin, and Christian Zwieb. “Systematic Site-Directed Mutagenesis of Human Protein SRP54: Interactions with Signal Recognition Particle RNA and Modes of Signal Peptide Recognition.” Biochemistry 41, no. 38 (September 24, 2002): 11362–11371. | ||
| - | |||
| - | # | ||
| - | # | ||
| - | |||
==Contributors== | ==Contributors== | ||
| - | + | Gregoire de Claviere and Hamelin Baptiste</StructureSection><!-- PLEASE DO NOT DELETE THIS TEMPLATE --> | |
{{Sandbox_Reserved_ESBS}} | {{Sandbox_Reserved_ESBS}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | ==Scènes à intégrer== | ||
| - | |||
| - | <scene name='56/568022/Hsrp54m_dimer/1'>hSRP54M Dimer</scene> | ||
| - | <scene name='56/568022/Hsrp54m_h1_to_h7/1'>hSRP54M and its 7 alpha helixes</scene> | ||
| - | <scene name='56/568022/Hsrp54m_core_and_h1/1'>Core and H1</scene> | ||
| - | <scene name='56/568022/Hsrp54m_conserved_residues/1'>Conserved residues</scene> | ||
| - | <scene name='56/568022/Hsrp54m_conserved_residues/2'>Conserved residues 2</scene> | ||
| - | <scene name='56/568022/Hsrp54m_core_and_h1/2'>Conserved residues 3</scene> | ||
| - | <scene name='56/568022/Hsrp54m_groove/1'>Hydrophobic groove</scene> | ||
| - | <scene name='56/568022/Hsrp54m_groove_with_h1/1'>Groove with H1</scene> | ||
| - | <scene name='56/568022/Hsrp54m_rna_bind/1'>Helixes involved in RNA binding</scene> | ||
| - | <scene name='56/568022/Hsrp54m_phe355_and_phe359/1'>Phe355 and Phe359</scene> | ||
| - | <scene name='56/568022/Hsrp54m_arg402_and_arg405/1'>Arg402 and Arg405</scene> | ||
| - | <scene name='56/568022/Hsrp54m_groovev2/1'>hydrophobic groovev2</scene> | ||
| - | <scene name='56/568022/Hsrp54m_rna_bindv2/1'>helices 4,5, 6 and 7</scene> | ||
| - | <scene name='56/568022/Hsrp54m_core_and_h1v2/1'>TextToBeDisplayed</scene> | ||
Revision as of 20:22, 8 January 2014
Human SRP54 M-domain
| |||||||||||
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |